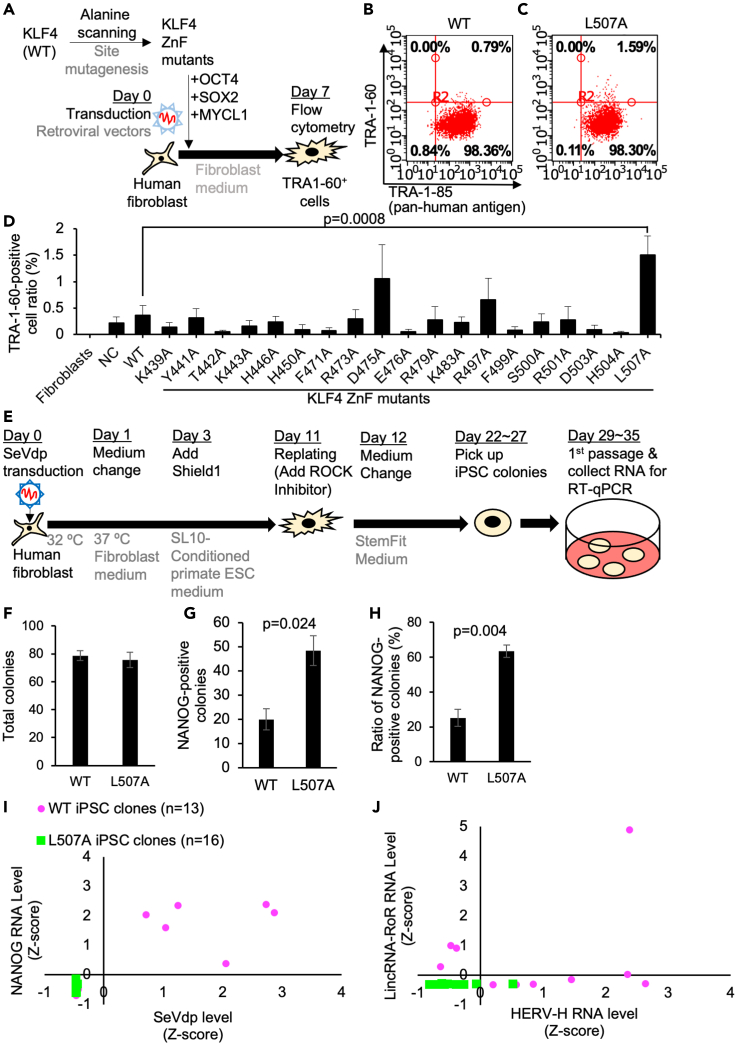
Figure 3.
The reprogramming efficiency and heterogeneity of human somatic cells using KLF4 ZnF alanine-scanned mutants
(A) Schematic of the reprogramming of human fibroblasts using alanine-scanned KLF4 ZnF mutants made by site mutagenesis of KLF4 wild-type (WT) retroviral vector together with OCT4, SOX2, and MYCL1 retroviral vectors.
(B and C) Typical data of flow cytometric analysis of the percentage of TRA1-60-positive cells in KLF4 alanine-scanned mutants 7 days after infection of reprogramming retrovirus vectors carrying KLF4 ZnF alanine-substituted mutants.
(D) Flow cytometric analysis of the percentage of TRA1-60-positive cells in KLF4 alanine-scanned mutants 7 days after infection of reprogramming retrovirus vectors carrying KLF4 ZnF alanine-substituted mutants. Results are mean and SE, n = 6. The p value was calculated with Dunnett's test between WT and L507A conditions.
(E) Schematic of the generation of human iPSCs using SeVdp carrying KLF4 or L507A mutant in feeder-free conditions. On day 3 after infection, the medium was changed to primate ESC medium-based conditioned medium which was collected from the culture of a feeder cell line. Because KLF4 is fused with a ligand-dependent destabilization domain (DD) at the N-terminus in this SeVdp, Shield1 to stabilize KLF4 protein in the ProteoTuner Shield system was added into one group until picking up iPSC colonies. Blasticidin S was added on day 3 after infection until day 7.
(F) Numbers of total colonies in each condition were counted by colony morphology on day 20. Results are mean and SE, n = 3.
(G) Numbers of NANOG-positive colonies in each condition were counted by immunocytochemistry. Results are mean and SE, n = 3. The p value calculated with the t test was shown based on the comparison between WT and L507A KLF4 conditions.
(H) The ratio of NANOG-positive colonies in each condition was calculated from numbers of total colonies and NANOG-positive colonies. Results are mean and SE, n = 3. The p value calculated with the t test was shown based on the comparison between WT and L507A KLF4 conditions.
(I and J) Variations of NANOG expression and residual SeVdp NP RNA amount (G) and HERV-H and lincRNA-RoR (H) in human iPSC clones generated with SeVdp carrying WT or L507A KLF4 were shown in scatter plots. Numbers (N) of the clones used in each condition are shown. Results are shown as Z-scores of all the samples.