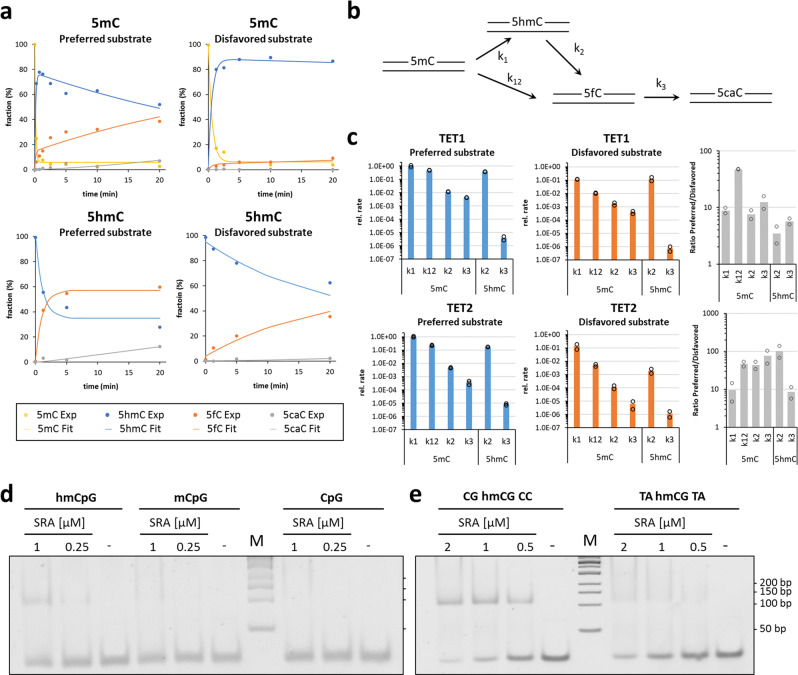
Fig. 3. Biochemical investigation of the flanking effect of TET1 and TET2 oxidation kinetics and 5hmC binding by UHRF2.
a Example of oxidation kinetics of synthetic double-stranded 30mer oligonucleotides containing one hemimethylated (5mC) or hemihydroxymethylated (5hmC) CpG site in different flanking context by TET2. Product appearance was detected by LC-MS. Enzyme concentrations of TET1 and TET2 were 1.6/2.0 µM for the favored substrates and 3.2/8 µM for the disfavored substrates. b Kinetic model used to analyze the data. c Summary of rate constants of oxidation of 5mC and 5hmC substrates by TET1 and TET2. Rates are given as relative values considering that in the reactions with the disfavored substrates more enzyme was used. Shown are averages and data points of two independent repeats. d Gel shift experiments with purified UHRF2 SRA domain and synthetic double-stranded 30mer oligonucleotides (0.5 µM) containing one hemihydroxymethylated CpG site (hmCpG), one hemimethylated (mCpG), and one unmodified CpG site in CGCGCC context. e Gel shift experiments with purified UHRF2 SRA domain and synthetic double-stranded 30mer oligonucleotides (0.5 µM) containing one hemihydroxymethylated CpG site in different flanking context.