Figure 6. Transposon promoters yield species-specific expression of evolutionarily conserved Cdk2ap1ΔN isoform.
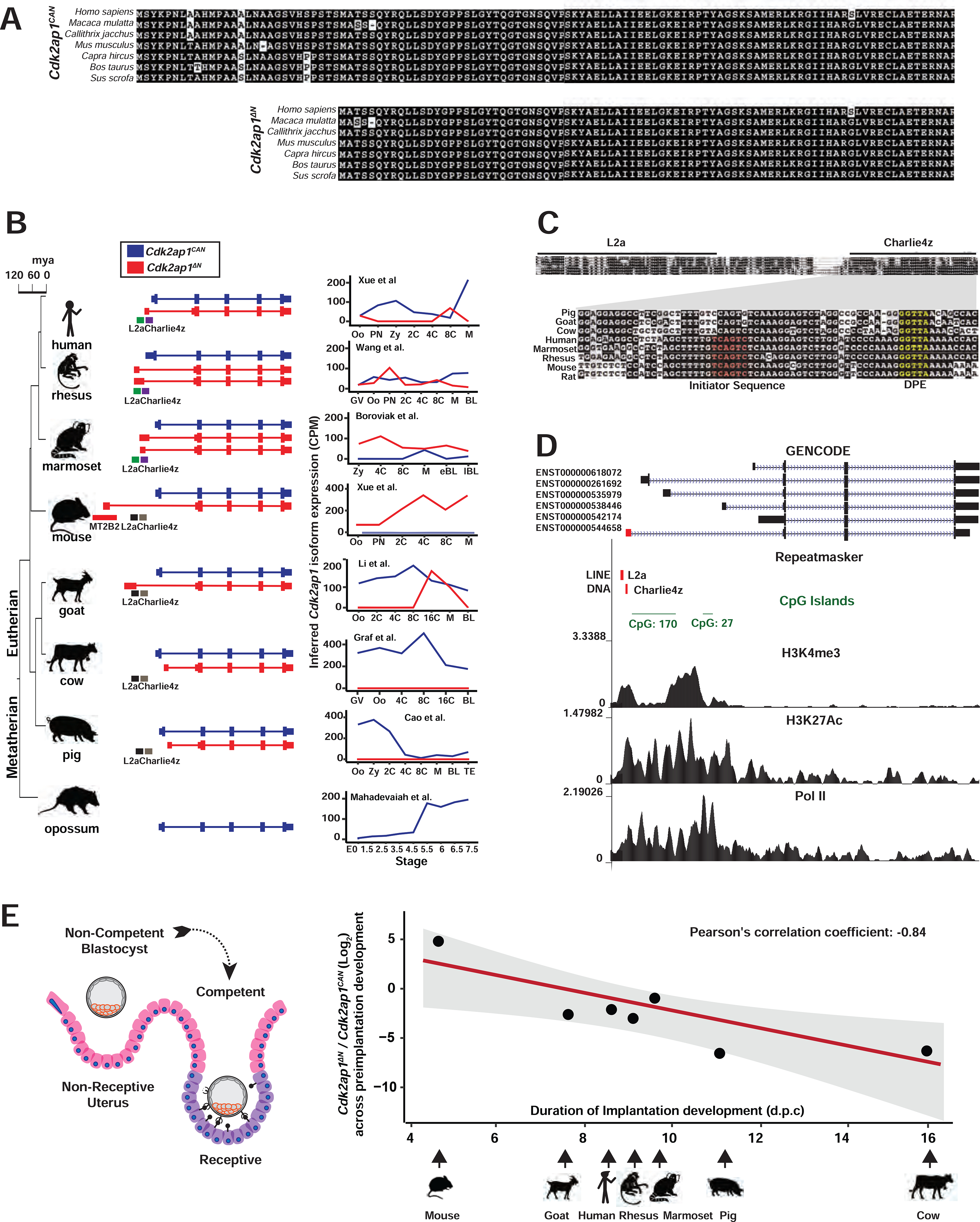
A. Alignment of Cdk2ap1CAN and Cdk2ap1ΔN isoforms across 8 mammals reveals strong evolutionary conservation in their protein sequences. B. Canonical Cdk2ap1 and Cdk2ap1ΔN exhibit species-specific differential expression in mammalian preimplantation embryos. Isoform specific expression of Cdk2ap1 in each species was determined by the total Cdk2ap1 expression and the ratio between isoform specific splicing junctions. C. In 8 mammals examined, the genomic regions containing the L2a/Charlie4z elements exhibit sequence conservation. The region between L2a and Charlie4z is the least conserved, with goat, pig and cattle harboring a small deletion, and rodents and primates exhibiting sequence variance. The Charlie4z element contains a predicted initiator sequence (red) and a DPE (Downstream Promoter Element, yellow), both implicating promoter functionality. D. The L2a/Charlie4z region acts as a bona fide CDK2AP1 promoter in human ESCs (Encode Consortium, 2012). Signatures of an active promoter (H3K4me3, H3K27Ac, and Pol II) in human ESCs are illustrated with ChIP-seq data from ENCODE and Roadmap Epigenomics project. E. The Cdk2ap1ΔN to Cdk2ap1CAN ratio is inversely correlated with the duration of preimplantation development in multiple mammals. The log2 ratio of Cdk2ap1ΔN to Cdk2ap1CAN, calculated based on the sum of normalized RNA-seq reads across isoform-specific junctions during preimplantation stages, is plotted against the duration of preimplantation development for each species. Pearson’s correlation coefficient between log2 (Cdk2ap1ΔN/Cdk2ap1CAN) and duration of preimplantation development equals to −0.84, ** P = 0.018, t = −3.5, df = 5; the P value was calculated as part of the Pearson’s product-moment correlation. See also Figure S6 and Tables S6 and S7.
