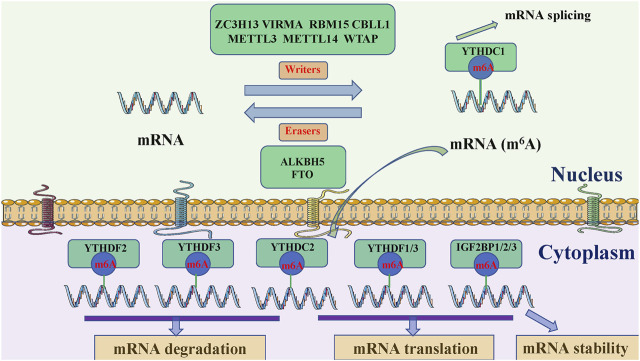
FIGURE 1.
Potential m6A methylation mechanisms in RNA. M6A methylation is catalyzed by the writer complex, including METTL3, METTL14, WTAP, VIRMA, RBM15, ZC3H13, and CBLL1. The demethylases, FTO and ALKBH5 remove m6A modifications. Reader proteins (YTHDC1, YTHDF2, YTHDF3, YTHDC2, YTHDF1/3, and IGF2BP1/2/3) recognize m6A and determine target RNA targets. METTL3, methyltransferase 3, N6-adenosine-methyltransferase complex catalytic subunit; METTL14, methyltransferase 14, N6-adenosine-methyltransferase subunit; WTAP, WT1 associated protein; VIRMA, vir like m6A methyltransferase associated; RBM15, RNA binding motif protein 15; ZC3H13, zinc finger CCCH-type containing 13; CBLL1, Cbl proto-oncogene like 1; FTO, FTO α-ketoglutarate dependent dioxygenase; ALKBH5, alkB homolog 5, RNA demethylase; YTHDC1/2, YTH domain containing 1/2; YTHDF1/2/3, YTH N6-methyladenosine RNA binding protein 1/2/3; IGF2BP1/2/3, insulin like growth factor 2 mRNA binding protein 1/2/3.