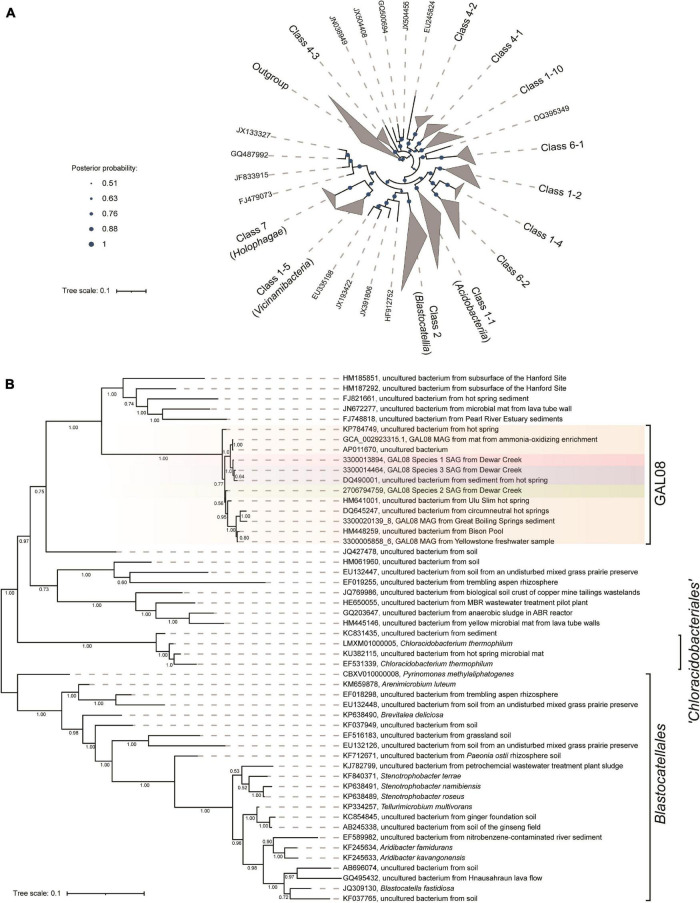
FIGURE 5.
(A) Bayesian 16S rRNA gene tree built using the ARB-Silva.de SSU rRNA gene reference database, release 138, showing the relationship of the major subgroups within the Acidobacteria. Nomenclature for the subgroups is used as suggested by Dedysh and Yilmaz (2018). The outgroup contains one 16S rRNA gene sequence from an isolate in each of 29 cultivated phyla. Confidence values, indicating posterior probabilities, are depicted as circles of different sizes. The scale bar represents 0.1 nucleotide substitutions per nucleotide position. (B) An expansion of the GAL08 within the Blastocatellia. All of the GAL08 sequences available in release 138 of the ARB-Silva database are included, as well as representative 16S rRNA gene sequences from each of the three species represented by SAGs and MAGs in this study. Two MAGs that contained 16S rRNA gene sequences from two other sites covered by the dataset of Nayfach et al. (2021) were also included. The sequences selected for Blastocatellia include all isolates and 1 to 2 sequences from each major clade available in release 138 of the ARB-Silva database. A posterior probability of 1.00 strongly supports the separation of GAL08 from the other members of the order Blastocatellales. The scale bar represents 0.1 nucleotide substitutions per nucleotide position. The list of accession numbers used in the construction of this tree is provided in Supplementary Table 1.