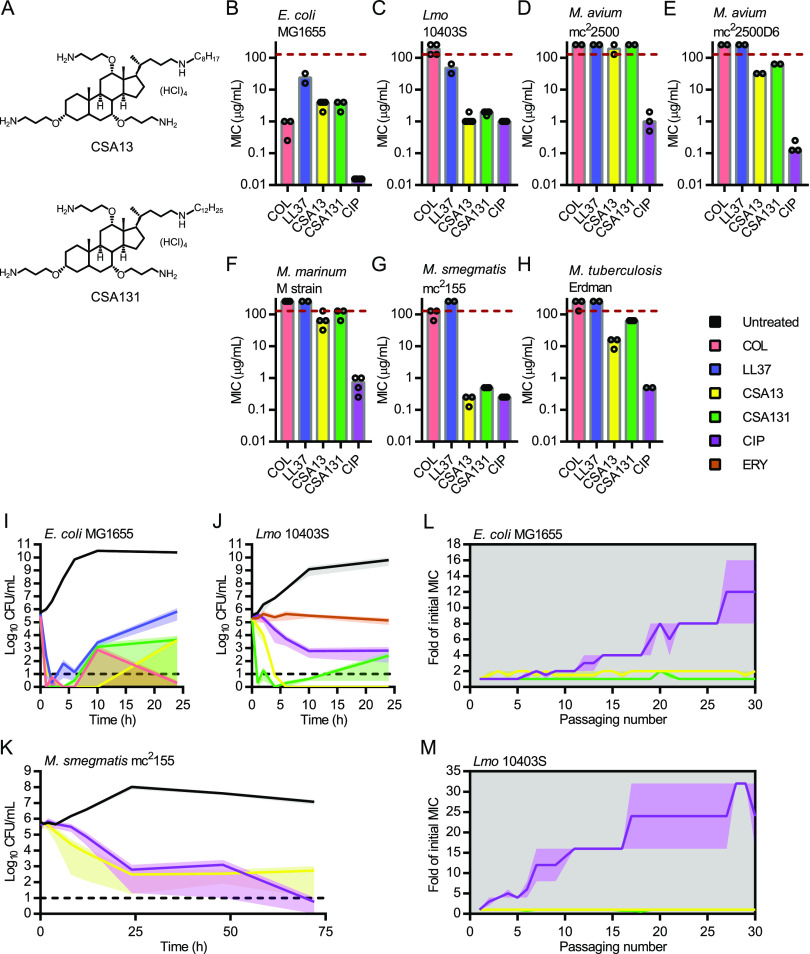
FIG 1.
Ceragenins kill phylogenetically diverse bacteria. (A) Structures of the ceragenins CSA13 and CSA131. MICs of colistin (COL), LL37, CSA13, CSA131, and ciprofloxacin (CIP) against E. coli MG1655 (B), L. monocytogenes (Lmo) 10403S (C), M. avium mc22500 (D), M. avium mc22500D6 (E), M. marinum M strain (F), M. smegmatis mc2155 (G) and M. tuberculosis Erdman (H). Dots and bars indicate results from independent experiments and median values, respectively. Time-kill experiments of E. coli MG1655 (I), L. monocytogenes 10403S (J), and M. smegmatis mc2155 (K) exposed to colistin (in pink), LL37 (in blue), CSA13 (in yellow), CSA131 (in green), ciprofloxacin (in purple) and/or erythromycin (ERY; in orange), a bacteriostatic antibiotic. Untreated samples are in black, and results are shown as means from two independent experiments. Shaded areas show standard error of the mean (SEM), and dotted lines indicate the limit of detection. Serial passages of E. coli (L) and L. monocytogenes (M) exposed to CSA13 (in yellow), CSA131 (in green), and ciprofloxacin (in purple). Bacteria were passaged daily in the presence of subinhibitory concentrations of antibiotics. Results are expressed as means and SEM from two independent experiments.