FIG 6.
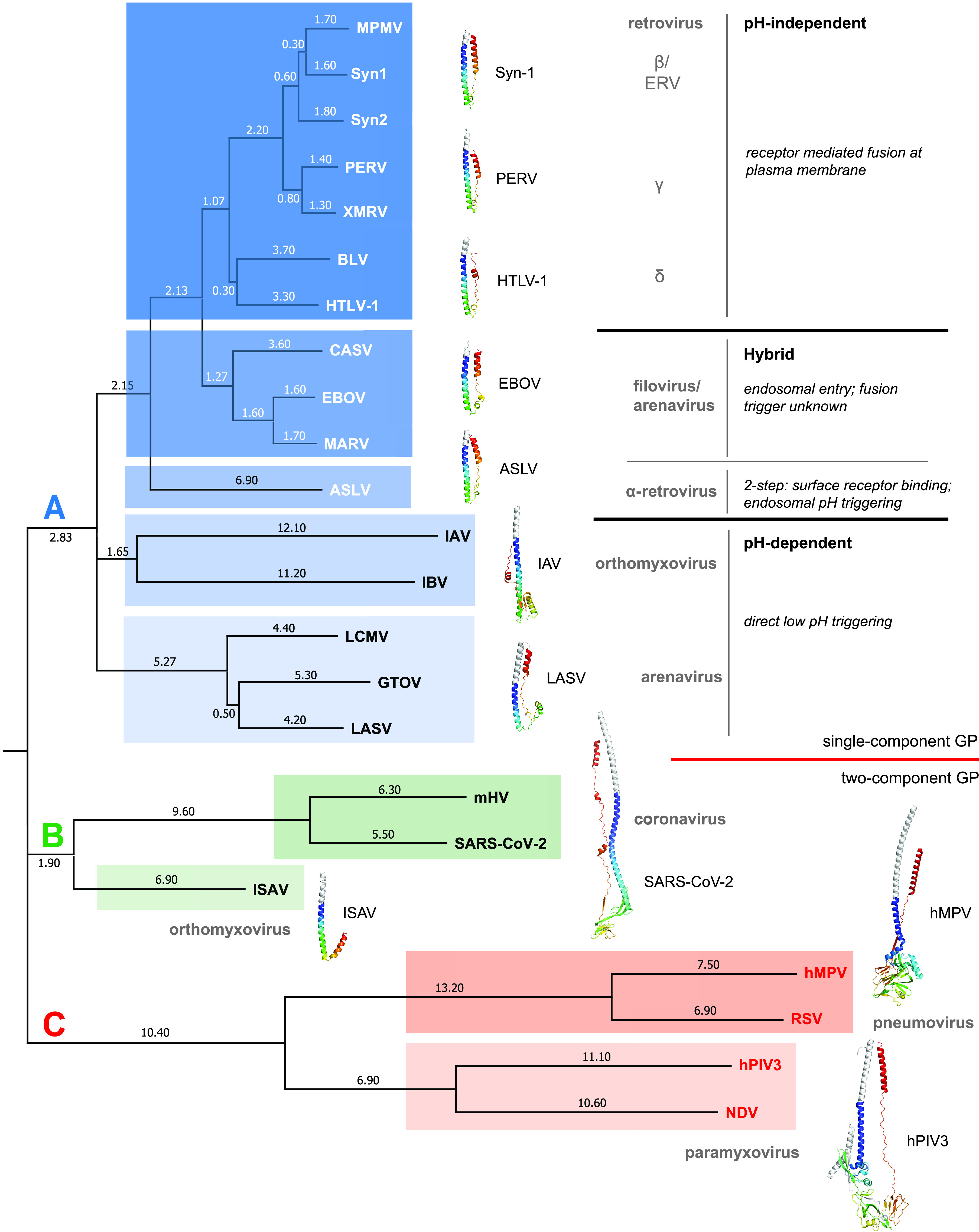
Structural dendrogram of class I postfusion subunits reveals clustering based on mechanism of viral entry. A structural dendrogram displaying the relationship between select class I viral postfusion structures as calculated by DALI (75–77). The evolutionary distances between structures are defined as the difference between the Z-score assigned to a structural pair and the sum of the Z-scores assigned to the self-alignment of each structure. Calculated evolutionary distances derived from the structural phylogenetic analysis are shown on the branch lines. Structures of representative viral fusion proteins from each family are displayed adjacent to the dendrogram. The distinct lineages are boxed and labeled in color. Viral classifications and fusion entry/triggering mechanisms are also annotated. Viruses are abbreviated as follows: ASLV, avian sarcoma leukosis virus; BLV, bovine leukemia virus; CASV, California Academy of Science virus; EBOV, Ebola virus; GTOV, Guanarito virus; hMV, human metapneumovirus; hPIV3, human parainfluenza virus 3; HTLV-1, human T-lymphotropic virus 1; IAV, influenza A virus; IBV, influenza B virus; ISAV, infectious salmon anemia virus; LASV, Lassa virus; LCMV, lymphocytic choriomeningitis virus; MARV, Marburg virus; MPMV, Mason-Pfizer monkey virus; mHV, murine hepatitis virus; NDV, Newcastle disease virus; RSV, respiratory syncytial virus; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; Syn-1, syncytin 1; Syn-2, syncytin 2; and XMRV, xenotropic murine leukemia virus-related virus.
