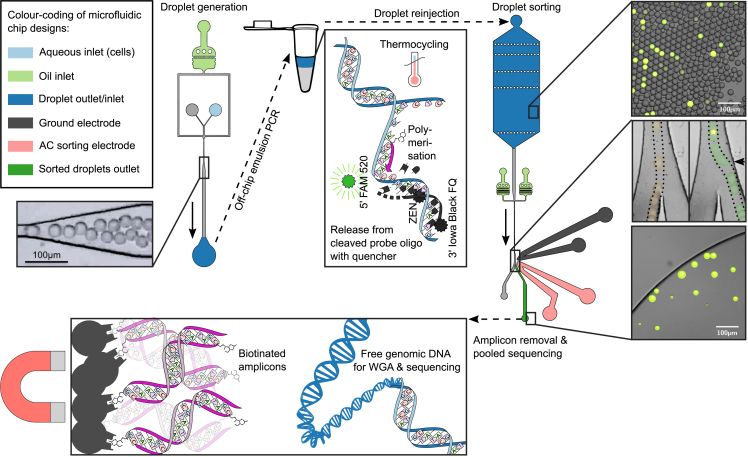
Figure 1.
Microfluidic workflow. Microfluidic chip designs, microscope images of their use, and schematics of molecular mechanisms
(Top left) DDDroplet generation chip and photo of 39 μm-droplet generation with cells and reagents in oil. The arrow indicates the flow direction. Top center: illustration of emulsion PCR in a thermocycler. If the target DNA sequence is present, it allows the binding of biotinylated primers and probes, thus the synthesis of corresponding amplicons. During strand extension, the TaqMan probes are cleaved, releasing fluorescent molecules.
(Top right) Droplet sorting chip design and microscopy images. The microscopy pictures on the right show an emulsion before sorting and during sorting and the positive droplet enrichment post sorting. Here, fluorescent droplets indicate the presence of the gut bacterium B. vulgatus inside the droplet. Multiple microscopy images of the droplet sorting junction at different time points were overlaid and colored to demonstrate droplet flow-traces. In the right image, a small black arrow indicates the location of the nearby sorting electrode, and a bright green spot indicates the upstream location of fluorescence detection. The scale bars correspond to 100 μm.
(Bottom) Illustration of the removal of abundant amplicons with biotin-binding Streptavidin beads (after pooling the sorted positive droplets) to purify genomic DNA for sequencing library preparation. Library preparation further involves whole-genome amplification (WGA) after binding adapter primers to the randomly broken genomic DNA fragments, followed by DNA fragment size selection.