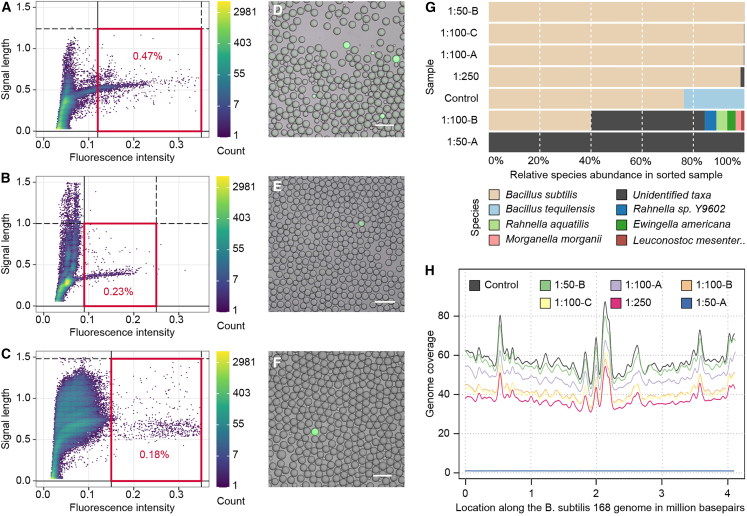
Figure 3.
Recovery of spike-in cells by droplet sorting. Fluorescence activated single-cell resolution droplet sorting of B. subtilis cells, which where spiked into stool samples at different ratios, of an emulsion with 1 in 4 droplets occupied by a cell. (A and D) About 1 B. subtilis cell in 50 microbiota cells, at 25% droplets occupied, means 1 positive droplet in ca. 200, ratio 0.005; (B and E) 1:100 cells, droplet ratio 0.0025; (C and F) 1:250 cells, droplet ratio 0.001
(A and B) Density plots of droplet sorting data with the applied sorting gates highlighted in red and with percentage of events within the gate. Sections of data are shown as these experiments were run over long time frames (ca. 10–20 h of sorting per sample), and gate boundaries had to be adjusted every few hours due to temperature changes and slightly varying droplet sizes. Repeated measurements yielded similar results.
(D–F) Microscopy overlay images of fluorescence and brightfield channel, showing positive droplets (containing B. subtilis culture) and negative droplets (empty or stool microbiota) post droplet PCR. The scale bars correspond to 100 μm.
(G) DNA sequencing data results of B. subtilis culture spiked into stool samples. The target read abundance is shown by sample. The unknown fraction (gray) could not be assigned to any bacterial genomes in our database, as further confirmed with Kraken assignments.
(H) Mapping of target DNA sequencing reads to the B. subtilis reference genome, showing the genome coverage of different samples.