Figure 4.
Enrichment and sequencing of endogenous target genome. Workflow summary of the precision genomics approach to enrich target endogenous species from its microbiome community with microfluidic droplets and summary of the two Bacteroides vulgatus genomes assembled in our experiments
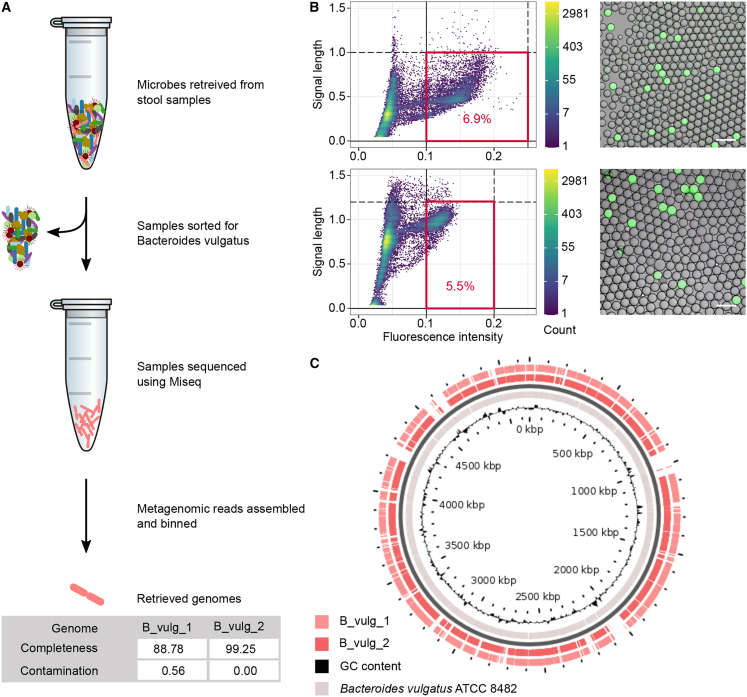
(A) Workflow schematic and genomes statistics.
(B) Two biological replicate experiments of fluorescence activated single-cell droplet sorting of endogenous Bacteroides vulgatus cells in stool samples, of an emulsion, with about 1 in 4 droplets occupied by a cell. (B, left) Density plots of droplet sorting data with the applied sorting gates highlighted in red and with percentage of events within the gate. Sections of data are shown. (B, right) Microscopy overlay images of fluorescence and brightfield channel, showing positive droplets (containing endogenous B. vulgatus) and negative droplets (empty or other stool microbiota) post droplet PCR. The scale bars correspond to 100 μm.
(C) The two assembled genomes were mapped to a reference genome using the blastx algorithm, illustrating (https://server.gview.ca/) the substantial genome coverage achieved here.