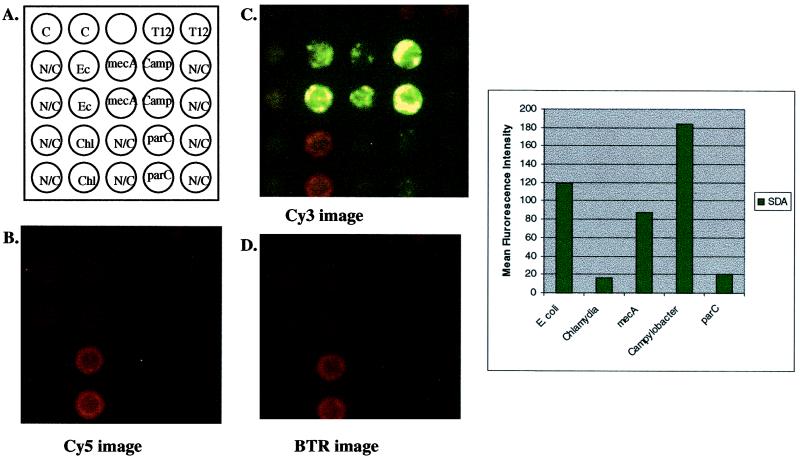
FIG. 5.
Multiplex amplification of E. coli gyrA, Staphylococcus mecA, Campylobacter gyrA, C. trachomatis cryptic plasmid, and E. coli parC. (A) Schematic representation of a multiplex (n = 5) amplification on the microelectronic chip array. Noncleavable (N/C) primers were included for each primer set to control for background hybridization of template DNA. Approximately 104 copies each of E. coli, methicillin-resistant S. aureus, fluoroquinolone-sensitive Campylobacter, and C. trachomatis cryptic plasmid genomic DNAs were combined and hybridized to each amplification primer set on the microelectronic chip array. Anchored SDA was initiated by the addition of enzymes and bumper primers for all systems. After amplification, reporters from all systems (wild-type and mutant reporters) were added and thermal stringency was applied at 29°C. Images were quantified for genotype identification of hybridized genomic DNA. (B) Cy5 fluorescent image after thermal stringency. Note that there is some bleed-through of the BTR signal seen in this image, due to overlapping emission of the BTR fluorophore at this wavelength. (C) Cy3 fluorescent image after thermal stringency. Note again the bleed-through of the BTR signal due to overlapping emission of the BTR fluorophore at this wavelength. (D) BTR image after thermal stringency. Quantification of signals after thermal stringency is shown in the graph.