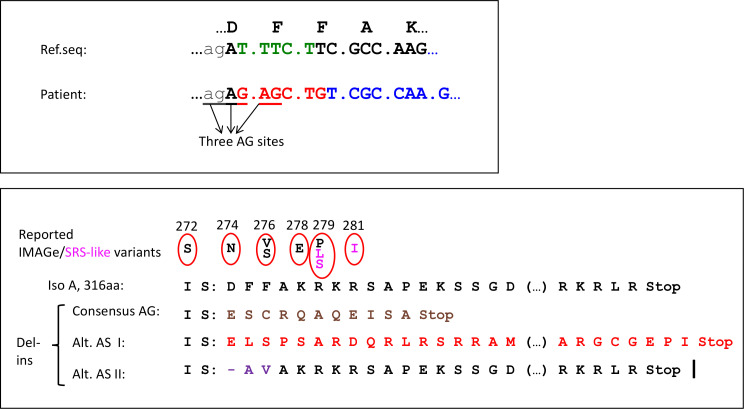
Figure 3.
Predicted impact of the delins variant c.822_826delinsGAGCTG on DNA and isoform A. Upper panel shows the CDKN1C intron 2/exon 3 junction with reference amino acids on top and the reference nucleotide sequence (ENST00000414822.7, CDKN1C-202) just below. Intron sequences are written in lower case letters and exon sequences in capital letters. The five deleted nucleotides are in green. The bottom line shows the mutated sequence with the six inserted nucleotides in red, the three adjacent AGs (potential acceptor sites/AS) underlined and the out-of-frame sequence marked in blue. The lower panel shows details of the predicted amino acid sequence and changes introduced by the delins variant in amino acid numbers according to protein isoform A (316aa—NP_000067.1). On top reported IMAGe (black) and Silver-Russel syndrome (SRS)–like variants (magenta) from literature with the corresponding amino acid change, then the reference protein product, followed by the mutated products using the consensus AG (p.Asp274GlufsTer12), the protein product if alternative acceptor site I is used (p.Asp274GlufsTer47) and the protein product if alternative acceptor site II is used (p.Asp274_276delinsAlaVal). Parentheses (…) represent 21 amino acids omitted from the figure.