Figure 1.
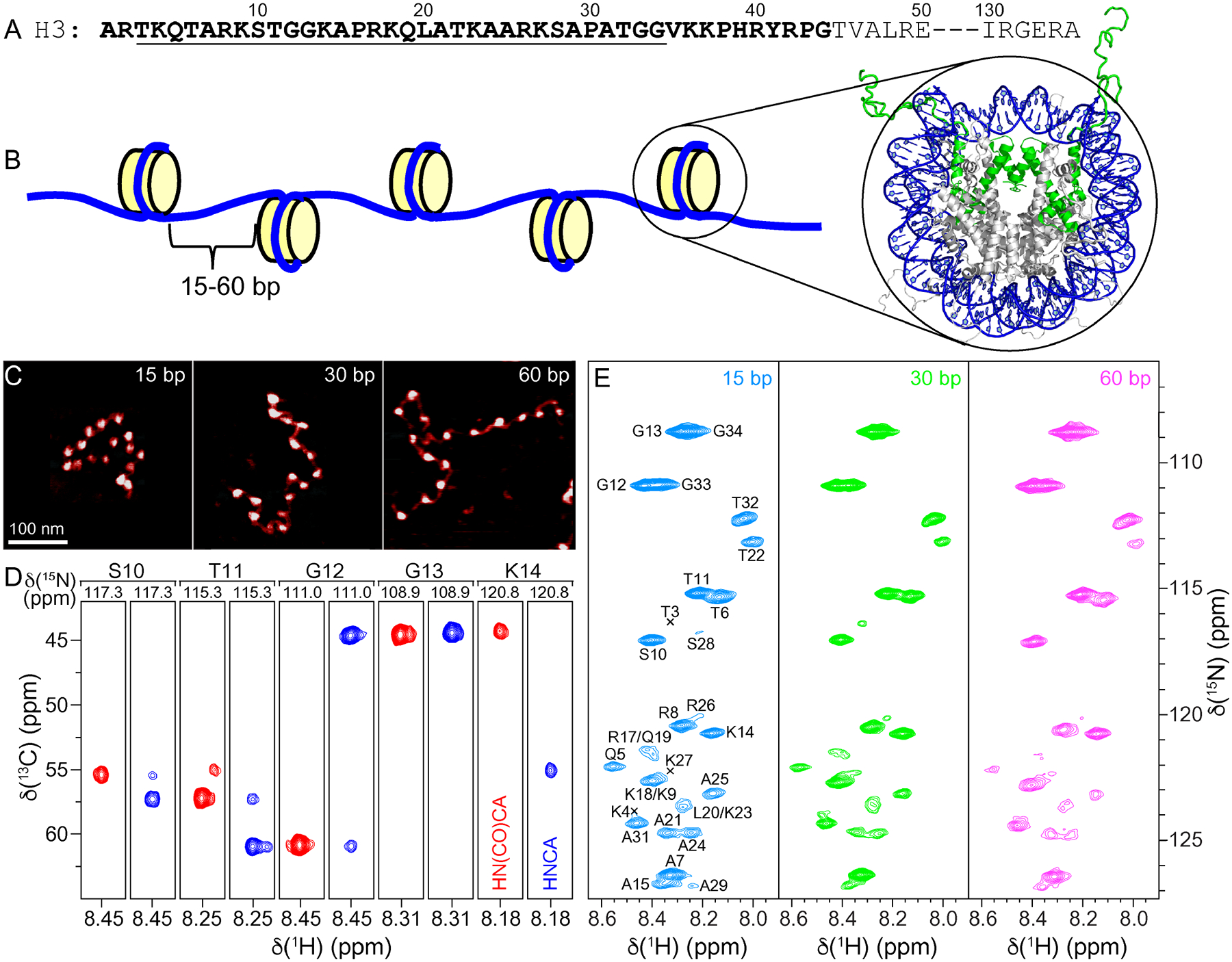
(A) Partial amino acid sequence of Xenopus laevis histone H3. Relatively unstructured residues based on the nucleosome core particle crystal structure are bold. Conformationally flexible residues detected in MAS NMR spectra of 16-mer nucleosome arrays in this study are underlined. (B) Schematic representation of the 16-mer nucleosome arrays with variable length (15, 30 or 60 bp) DNA linkers between nucleosome units. Also shown is the crystal structure of the nucleosome core particle (PDB entry 1KX5)32 with DNA and histone H3 colored blue and green, respectively, and histones H2A, H2B and H4 colored grey. (C) Representative AFM images of the 16-mer nucleosome arrays containing 15, 30 and 60 bp DNA linkers as indicated, showing the increase in internucleosome separation as a function of increasing linker DNA length. All images are shown on the same scale with the scale bar indicated in the leftmost panel. (D) Representative strips from 3D HNCA (blue contours) and HN(CO)CA (red contours) spectra of 16-mer nucleosome arrays with 15 bp DNA linkers reconstituted with 13C,15N-enriched histone H3, showing sequential connectivity for residues S10-K14. (E) 15N–1H HSQC spectra of 16-mer nucleosome arrays with 15 bp (blue), 30 bp (green), and 60 bp (magenta) DNA linkers, with the resonance assignments indicated. All spectra were recorded at 800 MHz 1H frequency, 10 kHz MAS rate, and sample temperature of ~35 °C.
