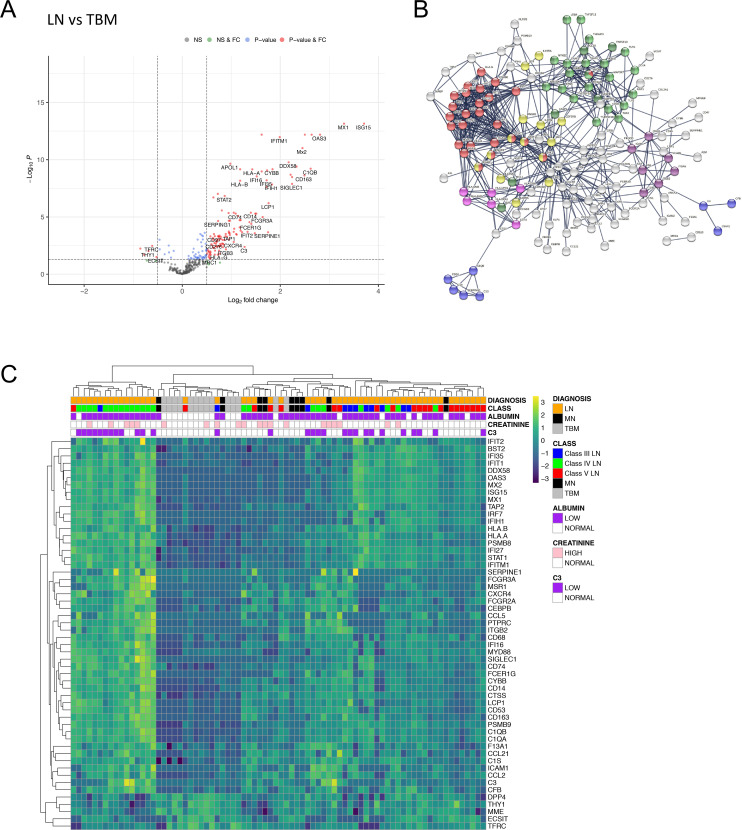
Figure 1.
DGE in LN compared with TBM disease. (A) Volcano plot depicting DGE of LN (n=55 biopsies) vs TBM disease (n=14 biopsies). Benjamini-Hochberg adjusted p=0.05 and log2 FC cut-off=0.5. (B) STRING interaction network for all 171 significant differentially expressed genes between LN and TBM (highest confidence score=0.9). Red nodes—type I IFN; blue nodes—complement cascade; pink nodes—MHC II; yellow nodes—JAK-STAT signalling; purple nodes—integrin binding; light green nodes—NF-κB+TIR domain; dark green nodes—NF-κB and apoptosis modulation. (C) Heatmap of all significantly increased differentially expressed transcripts with log2 FC ≥1 (n=49) and all significantly downregulated transcripts with log2 FC ≤−0.5 (n=5) identified in LN vs TBM, across the entire cohort (LN n=55 biopsies, MN n=9 biopsies, TBM n=14 biopsies). DGE, differential gene expression; FC, fold change; LN, lupus nephritis; MN, membranous nephropathy; NS, non-significant; TBM, thin basement membrane.