ABSTRACT
Human milk oligosaccharides (HMOs), which are natural bifidogenic prebiotics, were recently commercialized to fortify formula milk. However, HMO assimilation phenotypes of bifidobacteria vary by species and strain, which has not been fully linked to strain genotype. We have recently shown that specialized uptake systems, particularly for the internalization of major HMOs (fucosyllactose [FL]), are associated with the formation of a Bifidobacterium-rich gut microbial community. Phylogenetic analysis revealed that FL transporters have diversified into two clades harboring four clusters within the Bifidobacterium genus, but the underpinning functional diversity associated with this divergence remains underexplored. In this study, we examined the HMO consumption phenotypes of two bifidobacterial species, Bifidobacterium catenulatum subsp. kashiwanohense and Bifidobacterium pseudocatenulatum, both of which possess FL-binding proteins that belong to phylogenetic clusters with unknown specificities. Growth assays, heterologous gene expression experiments, and HMO consumption analyses showed that the FL transporter type from B. catenulatum subsp. kashiwanohense JCM 15439T conferred a novel HMO uptake pattern that includes complex fucosylated HMOs (lacto-N-fucopentaose II and lacto-N-difucohexaose I/II). Further genomic landscape analyses of FL transporter-positive bifidobacterial strains revealed that the H-antigen- or Lewis antigen-specific fucosidase gene(s) and FL transporter specificities were largely aligned. These results suggest that bifidobacteria have acquired FL transporters along with the corresponding gene sets necessary to utilize the imported HMOs. Our results provide insight into the species- and strain-dependent adaptation strategies of bifidobacteria in HMO-rich environments.
IMPORTANCE The gut of breastfed infants is generally dominated by health-promoting bifidobacteria. Human milk oligosaccharides (HMOs) from breast milk selectively promote the growth of specific taxa such as bifidobacteria, thus forming an HMO-mediated host-microbe symbiosis. While the coevolution of humans and bifidobacteria has been proposed, the underpinning adaptive strategies employed by bifidobacteria require further research. Here, we analyzed the divergence of the critical fucosyllactose (FL) HMO transporter within Bifidobacterium. We have shown that the diversification of the solute-binding proteins of the FL transporter led to uptake specificities of fucosylated sugars ranging from simple trisaccharides to complex hexasaccharides. This transporter and the congruent acquisition of the necessary intracellular enzymes allow bifidobacteria to consume different types of HMOs in a predictable and strain-dependent manner. These findings explain the adaptation and proliferation of bifidobacteria in the competitive and HMO-rich infant gut environment and enable accurate specificity annotation of transporters from metagenomic data.
KEYWORDS: Bifidobacterium, human milk oligosaccharides (HMOs), fucosyllactose transporter, fucosidase
INTRODUCTION
Human milk oligosaccharides (HMOs) are a diverse group of oligosaccharides (degree of polymerization of ≥3) with more than 200 different structures and are the third most abundant solid component in breast milk (1, 2). HMOs are composed of five monosaccharides (fucose [Fuc], galactose [Gal], glucose [Glc], N-acetylglucosamine [GlcNAc], and N-acetylneuraminic acid [sialic acid]) and are synthesized through the elongation and/or modification of lactose (Lac) in the mammary gland (see Fig. S1 in the supplemental material for HMO structures). The majority of HMOs are fucosylated, with the most abundant being the trisaccharides 2′-fucosyllactose (2′-FL) and 3-fucosyllactose (3-FL) (3). Fucosylation is catalyzed by fucosyltransferase 2 (FUT2) and fucosyltransferase 3 (FUT3) (1). Depending on the geographic region, the proportion of secretor mothers (FUT2+/+ or FUT2+/−) can range from 65% to 98%, which affects the composition of HMOs in breast milk (4). For example, the milk of secretor mothers generally contains 2′-FL as the most abundant HMO species, while nonsecretor mothers produce more lacto-N-fucopentaoses (LNFPs) II and III, which possess α-1,4- and α-1,3-fucosyl substitutions, respectively, to compensate for the absence of 2′-FL (4). Despite the high energy expenditure for the mother in their production, HMOs provide no nutritional value to the infant as they are resistant to pancreatic digestion (5). Rather, they function as selective substrates that promote the growth of HMO-utilizing taxa in the gut, such as bifidobacteria (6–8), which are more dominant in breastfed infants than in formula-fed infants (9). Consequently, there has been an increased effort to fortify commercial infant formula with HMOs. Previous studies have shown that bifidobacteria possess the extensive enzymatic machinery to metabolize HMOs (10–12), reflecting their adaptation to the breastfed infant gut environment (13, 14). However, HMO consumption ability can vary at the strain and even at the homolog levels (12), and the variation in their phenotypes has not been fully linked to their genotypes. Therefore, insights into HMO assimilation and adaptation strategies employed by Bifidobacterium strains require further research.
Several other gut microbes possess enzymes to break down HMOs (e.g., fucosidases) and display an extent of growth on HMOs in pure cultures, e.g., members of Bacteroides (15) and Akkermansia muciniphila (16). These taxonomic groups, however, lack the unique and efficient HMO uptake systems of bifidobacteria. HMOs are imported by bifidobacteria using ATP-binding cassette (ABC) transporters (one notable exception is Bifidobacterium bifidum, which extracellularly degrades HMOs [11, 17]). The solute-binding protein (SBP) of an ABC transporter mediates the high-affinity capture of ligands and initiates their translocation, after which depolymerization in the cytoplasm is catalyzed by a series of glycoside hydrolases (GHs) (6, 18–22). As the majority of HMOs are fucosylated, the characterization of fucosyllactose (FL) transporters is critical for revealing how bifidobacteria have adapted to the infant gut environment. In a previous study (20), we characterized two ABC transporters that import fucosylated HMOs (FL transporter 1 [SBP name, FL1-BP] and FL transporter 2 [SBP name, FL2-BP]) found in Bifidobacterium longum subsp. infantis. The two transporters have identical permease subunits, as opposed to their divergent SBPs (60% shared identity) that confer different specificities: while the specificity of FL transporter 1 is limited to 2′-FL and 3-FL, FL transporter 2 imports a wider variety of fucosylated HMOs (2′-FL, 3-FL, lactodifucotetraose [LDFT], and LNFP I). Furthermore, the presence of FL1-BP and FL2-BP homologs in fecal samples was strongly associated with a Bifidobacterium-rich microbiota in breastfed infants. Taken together, these findings suggest that these FL transporters represent an adaptive strategy employed by bifidobacteria to proliferate in the infant gut.
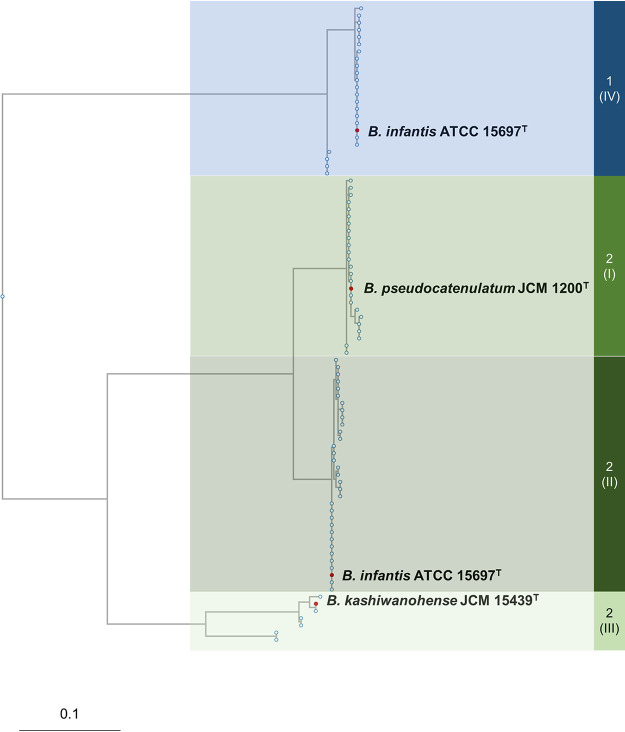
Our previous phylogenetic analysis of the FL1-BP and FL2-BP homologs, which are exclusively distributed in bifidobacterial genomes, also revealed the presence of four clades comprising cluster 1-IV (FL1-BP) and clusters 2-I, 2-II, and 2-III (FL2-BP) (Fig. 1) (20). While previous studies have extensively focused on FL transporters belonging to B. infantis ATCC 15697T (clusters 1-IV and 2-II), clusters 2-I and 2-III warrant further research (6, 19, 20, 23, 24). Of the two clusters, cluster 2-III displays the lowest identity to cluster 2-II (71%). Bifidobacterium catenulatum subsp. kashiwanohense JCM 15439T, which harbors an FL-BP homolog from cluster 2-III, was first isolated from infant feces (25), and its average relative abundance in infant feces can reach as high as 22% (26). James et al. (24) showed through growth assays that B. kashiwanohense APCKJ1 is capable of utilizing 2′-FL and 3-FL through the use of its FL transporter (cluster 2-III), but heterologous transporter expression experiments by our group (20) showed that the transporter also imports LDFT and LNFP I. The ability to consume fucosyllactose is well conserved within the B. kashiwanohense species (27), but the substrate specificity of FL transporter cluster 2-III for other HMO molecules has not yet been described.
FIG 1.
Phylogenetic analysis of FL transporter SBPs. A phylogenetic tree was constructed based on the amino acid sequence of the FL transporter 2 SBP (Blon_2202) found in B. infantis ATCC 15697T. Eighty-nine homologs from 69 strains that share ≥60% identity are shown, and four different clusters were identified, as expected from our previous study (20). Representative strains of each cluster are shown. Note that B. infantis ATCC 15697T is shown twice as several B. infantis strains harbor transporters from both clusters 2-II and 1-IV. Also, see Table S1 in the supplemental material for more detail.
In this study, we describe the physiological relevance of clusters 2-I and 2-III of FL transporter 2 for HMO assimilation through growth assays, heterologous gene expression experiments, and HMO consumption analyses. Our results showed that FL transporter 2 (cluster 2-III) found in B. kashiwanohense possesses novel substrate specificity toward fucosylated HMOs with Lewis a and b antigens [Galβ1,3(Fucα1,4)GlcNAc and Fucα1,2-Galβ1,3(Fucα1,4)GlcNAc, respectively]. Further gene landscape analysis of known bifidobacterial genomes revealed that the presence of fucosidase genes complemented the substrate specificity of FL transporters. The results of our study provide further evidence to suggest that the diversification of the FL transporter is a genomic signature that reveals the adaptation of bifidobacteria to HMO environments.
RESULTS
Diversification of FL-BP homologs within bifidobacterial species.
We performed an updated analysis with tBLASTn of FL2-BP (20) homologs to include newly deposited genome sequences in the National Center for Biotechnology Information (NCBI) genomic database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Fig. 1). FL2-BP found in B. infantis ATCC 15697T (Blon_2202 [GenBank accession no. CP001095.1]) was used as a query sequence. Homologs (≥60% amino acid sequence identity) were found only in bifidobacterial genomes. As shown in Fig. 1, the homologs formed two phylogenetic clades (clades 1 and 2), which is consistent with previous work, and the clades were further categorized into four clusters with distinctive ligand-binding residues (20). The clusters showed various degrees of identity to Blon_2202: cluster 2-II showed 98 to 100% identity, represented by B. infantis ATCC 15697T; cluster 2-I showed 90 to 92% identity, represented by Bifidobacterium pseudocatenulatum JCM 1200T; cluster 2-III showed 71 to 72% identity, represented by B. kashiwanohense JCM 15439T; and cluster 1-IV showed 61% identity, represented by B. infantis ATCC 15697T and Bifidobacterium breve lw01 (Fig. 1; see also Table S1 in the supplemental material). Of note, the sequences of the corresponding permease subunits of the FL transporters were conserved across the four clusters (≥88% identity), suggesting that the specificities of FL transporters are effectuated by the diversification of the SBPs.
A novel and versatile transporter specificity toward mono- and difucosylated HMOs in B. kashiwanohense.
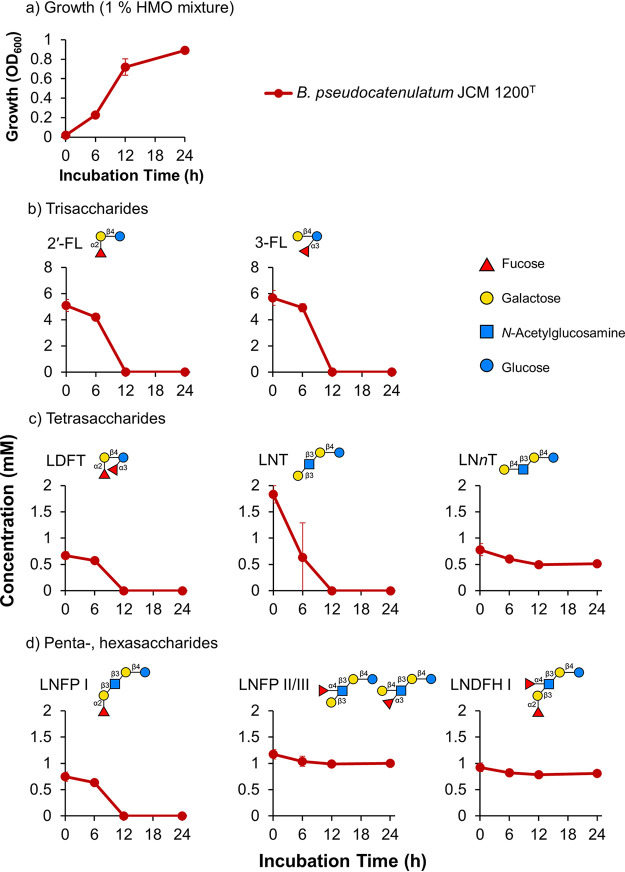
To examine the HMO consumption patterns of bifidobacteria with FL transporters (clusters 2-I and 2-III), we performed growth assays in a medium containing 1% HMOs as the sole carbon source. As shown in Fig. 2, we first examined B. pseudocatenulatum harboring the cluster 2-I transporter. B. pseudocatenulatum grew well in HMO-supplemented medium (optical density at 600 nm [OD600] of >0.8) (Fig. 2a). 2′-FL, 3-FL, LDFT, lacto-N-tetraose (LNT), and LNFP I were depleted from the culture supernatant during growth (Fig. 2b to d; Fig. S2a). In contrast, the strain did not assimilate lacto-N-neo-tetraose (LNnT), LNFP II/III, or lacto-N-difucohexaose (LNDFH) I (Fig. 2c and d). As expected from the high amino acid similarity (∼90%) with cluster 2-II, these results strongly suggested that cluster 2-I of B. pseudocatenulatum conferred the same HMO assimilation phenotype as cluster 2-II (represented by FL2-BP of B. infantis ATCC 15697T).
FIG 2.
Growth and HMO consumption behavior of B. pseudocatenulatum JCM 1200T. B. pseudocatenulatum JCM 1200T was grown in MRS-CS medium containing 1% (wt/vol) HMOs as a sole carbon source. (a) Growth was monitored by measuring the OD600. Additionally, the supernatant of the culture medium was sampled at the indicated time points and analyzed using HPLC. (b to d) Concentrations of trisaccharides (b), tetrasaccharides (c), and penta- and hexasaccharides (d) remaining in the medium. Data are means ± standard deviations from biological triplicates. Note that LNTri-II was not detected, and LNDFH II was not quantified as no changes were observed over time (see Fig. S2a in the supplemental material).
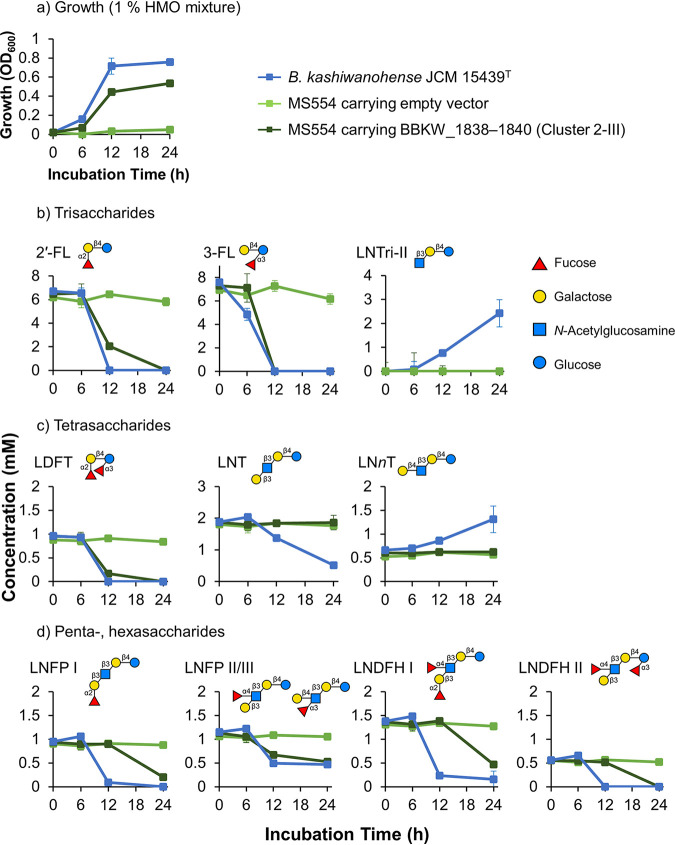
We then examined the HMO consumption pattern of B. kashiwanohense with the cluster 2-III transporter, as shown in Fig. 3. B. kashiwanohense grew well in HMO-supplemented medium (OD600 of >0.7) (Fig. 3a) and displayed a novel HMO assimilation phenotype, as it consumed LNFP II/III, LNDFH I, and LNDFH II in addition to 2′-FL, 3-FL, LDFT, and LNFP I (Fig. 3b to d; Fig. S2b). However, it consumed neither LNT nor LNnT (Fig. 3c). Furthermore, high-performance liquid chromatography (HPLC) analysis of the culture supernatant showed an accumulation of a peak predicted to be the trisaccharide lacto-N-triose II (LNTri-II) (retention time of 35 to 37 min) (Fig. S2b). We verified that the peak was LNTri-II [GlcNAc(β1,3)Gal(β1,4)Glc] through enzymatic analysis (Fig. S3). We note that although B. kashiwanohense possesses a homolog of an LNTri-II-degrading enzyme (GH20 β-1,3-N-acetylglucosaminidase; 59% identity to Blon_0459 [28]), LNTri-II accumulation was observed, and the mechanism remains unclear. Due to this novel fucosylated HMO assimilation phenotype, we focused on cluster 2-III to further examine its ability to import HMOs. To determine whether the assimilation of HMOs was attributable to the transporter, we performed growth assays using previously constructed heterologous expression strains. The strain, a B. longum JCM 31944 ΔlnbX ΔgltA strain that heterologously expresses the genes encoding intracellular exo-glycosidases for HMO depolymerization (MS554 strain), was previously transformed with a plasmid expressing the FL transporter belonging to cluster 2-III (BBKW_1838 to BBKW_1840 [BBKW_1838–1840; GenBank accession no. AP012327.1]) or an empty vector as a negative control (20). Growth assays revealed that the strain carrying BBKW_1838–1840 grew moderately well (OD600 of >0.5) in HMO-supplemented medium, in contrast to the control MS554 strain carrying an empty vector (OD600 of <0.1) (Fig. 3a). Furthermore, MS554 carrying BBKW_1838–1840 displayed HMO consumption behavior similar to that of B. kashiwanohense, in which 2′-FL, 3-FL, LDFT, and LNFP II/III were consumed first, followed by LNFP I, LNDFH I, and LNDFH II (Fig. 3b to d; Fig. S2c and d). Unlike B. kashiwanohense, accumulation of LNTri-II was not observed in the culture supernatant of MS554 carrying BBKW_1838–1840.
FIG 3.
A novel transporter specificity capable of importing complex fucosylated HMOs. Strains were grown in MRS-CS medium containing 1% (wt/vol) HMOs as a sole carbon source. (a) Growth was monitored by measuring the OD600. Additionally, the supernatant of the culture medium was sampled at the indicated time points and analyzed using HPLC. (b to d) Concentrations of trisaccharides (b), tetrasaccharides (c), and penta- and hexasaccharides (d) remaining in the medium. Data are means ± standard deviations from biological triplicates.
Differentiation of LNFPs II and III was not possible through HPLC analysis, as they are detected as a combined peak. To further determine the specificity of cluster 2-III, we performed additional enzymatic assays with the fraction containing the combined LNFP II/III peak (retention time of 55 to 57 min) (Fig. S2b and d) from the supernatant at 24 h, as indicated in Fig. S4. A reaction with AfcB (29), an α-1,3/4-l-fucosidase, was predicted to liberate LNT if LNFP II, which contains a Lewis a antigen, remained in the medium (Fig. S4a). On the other hand, it would release LNnT if LNFP III, which contains a Lewis x antigen, was left in the medium. HPLC analysis after enzymatic treatment showed that LNnT was liberated, indicating that LNFP III remained in the medium, and LNFP II was consumed by the cluster 2-III transporter (Fig. S4b). FL transporter 2 (cluster 2-III) from B. kashiwanohense JCM 15439T can assimilate LNT structure oligosaccharides with H-antigens (Fucα1,2Gal) and Lewis a [Galβ1,3(Fucα1,4)GlcNAc] and Lewis b [Fucα1,2Galβ1,3(Fucα1,4)GlcNAc] antigens and is also specific for mono- and difucosylated lactose molecules. These results indicated that cluster 2-III of FL transporter 2 was distinguished by the uptake of the broadest range of fucosylated HMOs (2′-FL, 3-FL, LDFT, LNFP I, LNFP II, LNDFH I, and LNDFH II) among the FL transporter clusters.
The presence of fucosidase genes corresponds to the substrate specificity of each FL transporter type.
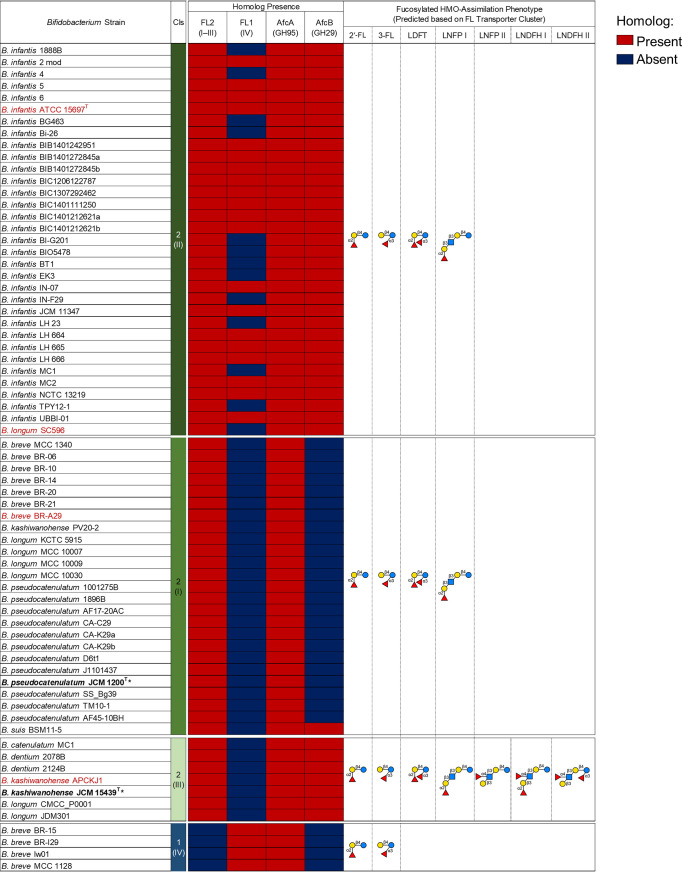
We performed gene landscaping analysis of available genomes of FL transporter-positive bifidobacterial strains (Fig. 1; Table S1), which revealed differential patterns in the presence of fucosidase genes (Fig. 4; Fig. S5). As described above, the transporter types can vary by strain. For example, the majority of B. infantis strains possess both cluster 2-II and 1-IV transporter proteins. As for B. kashiwanohense strains, two possess the transporter assigned to cluster 2-III, and one possesses the transporter from cluster 2-I. As summarized in Fig. 4, further analysis showed that all strains included in the analysis possessed putative H-antigen-specific intracellular 1,2-α-l-fucosidases of GH95 (homologs of AfcA from B. infantis [21]), but there was an apparent absence of Lewis antigen-specific intracellular 1,3/4-α-l-fucosidases of GH29 (homologs of AfcB from B. infantis [21]) in strains harboring FL transporters from clusters 1-IV and 2-I. The overall genomic organizations at the Fuc and FL utilization loci among transporter types are similar (Fig. S5). However, through identifying the substrate specificities of previously uncharacterized FL transporter types, we found that the presence of AfcA and/or AfcB corresponds to the substrate specificity of each FL transporter type. Of note, B. infantis strains possess AfcB, although AfcB is not required for the degradation of HMOs imported by cluster 1-IV and/or 2-II transporter types; however, B. infantis strains are known to import many types of fucosylated HMOs through other unidentified transporters (12, 30), for which AfcB most likely becomes relevant.
FIG 4.
Correlation of fucosidase and FL transporter specificities in Bifidobacterium strains. The distributions of fucosidase homologs among Bifidobacterium strains harboring FL transporter homologs are shown. Red indicates that a homolog is present (identity of >70%), and dark blue indicates that the homolog is absent from the strain. B. longum strains without subspecies identification were excluded from the analysis. The HMO assimilation phenotype based on the FL transporter type is shown to the right. Functions of transporters belonging to the strains indicated in red have previously been explored to a different resolution (6, 19, 20, 22, 24). The strains used in this study are indicated in boldface type with asterisks. Cls, clusters.
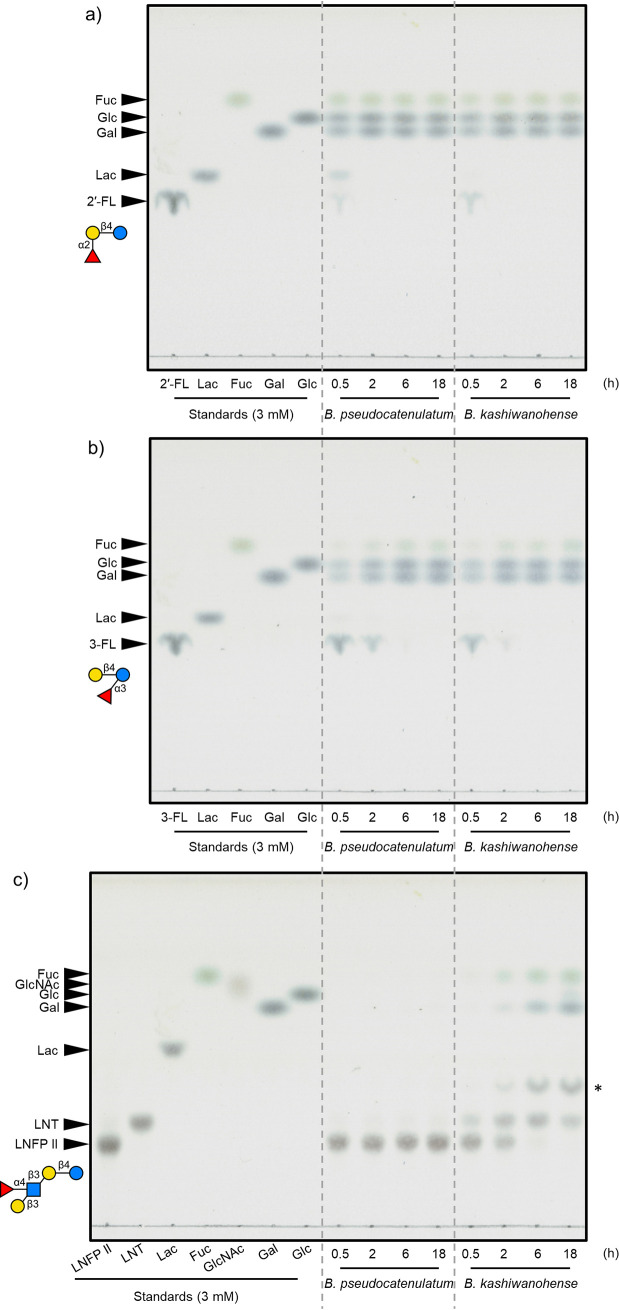
As shown in Fig. 5, we then incubated cell extracts with 2′-FL, 3-FL, and LNFP II to confirm the presence of AfcA and AfcB activity within bifidobacterial strains harboring either cluster 2-I (B. pseudocatenulatum JCM 1200T) or cluster 2-III (B. kashiwanohense JCM 15439T). AfcA preferentially acts on α-1,2 linkages and also on 3-FL (α-1,3 linkage) but to a significantly lesser degree (17, 21, 24). On the other hand, AfcB exclusively acts on α-1,3/4 linkages between Fuc and GlcNAc/Glc with branched Gal residues (21, 24, 31). Cell extracts from B. pseudocatenulatum almost completely degraded 2′-FL within 2 h, while 3-FL degradation was delayed (Fig. 5a and b). However, no activity was observed on LNFP II, supporting the absence of AfcB in B. pseudocatenulatum (Fig. 5c). B. kashiwanohense, on the other hand, degraded all three sugars, confirming the presence of both AfcA and AfcB (Fig. 5a to c). We also note that when cell extracts of B. kashiwanohense were incubated with LNFP II, there was an accumulation of what appears to be LNTri-II (Fig. 5c). These results indicate that the presence of AfcB is necessary for the degradation of a wider range of fucosylated HMOs and that the conservation of intracellular fucosidases corresponds well to the substrate specificities of FL transporters.
FIG 5.
Fucosylated oligosaccharide degradation by cell extracts from bifidobacterial strains with different transporter specificities. Cell extracts (protein concentration of 64.1 μg/mL) from B. pseudocatenulatum JCM 1200T or B. kashiwanohense JCM 15439T were incubated at 37°C on 3 mM 2′-FL (a), 3-FL (b), or LNFP II (c). The samples were collected at the indicated time points and analyzed by thin-layer chromatography as previously described (40). The spot indicated by the asterisk is predicted to be LNTri-II. Experiments were conducted in biological duplicates, and representative results are shown.
DISCUSSION
The majority of HMOs in breast milk are fucosylated (2–4), and their utilization by bifidobacteria represents a recently growing interest, especially in the fortification of formula milk. The ability to utilize fucosylated HMOs has been suggested to be a strategy employed by bifidobacteria to adapt to the infant gut environment, and the gene loci of FL utilization have been previously reported (6, 12, 20, 22, 24) (see Fig. S5 in the supplemental material). Despite the clear homology of SBPs associated with FL transporters (identity of ≥60%), these proteins segregate into two main phylogenetic clades harboring distinct clusters (clusters 2-I, 2-II, 2-III, and 1-IV) (Fig. 1; Table S1). The importance of the transporters in the infant gut in promoting a bifidobacterial community has been demonstrated recently (6, 20). However, differences in substrate specificities had not been clarified, as the abilities of the transporters to confer HMO assimilation phenotypes were assumed to be similar among clusters assigned to each clade (especially clusters 2-I, 2-II, and 2-III) (6, 19, 20, 23, 24). In addition to FL transporters, a common feature of FL-utilizing bifidobacterial strains is the presence of intracellular 1,2-α-l-fucosidases (AfcA) (Fig. 4; Fig. S5). While AfcA is sufficient for the metabolism of most fucosylated HMOs (21, 24), some strains also possess an intracellular 1,3/4-α-l-fucosidase (AfcB). However, there were no evident patterns in the presence and distribution of intracellular AfcB, and its physiological role had been unclear. Here, our analysis now clearly shows that the FL transporter has diversified to display various substrate specificities and that the difference in substrate specificities can explain the occurrence patterns of intracellular AfcA and AfcB in bifidobacteria. Such diversification highlights an important adaptation employed by bifidobacterial strains to utilize fucosylated HMOs, and our study provides molecular insight into strategies that bifidobacteria may have developed in response to the infant gut environment.
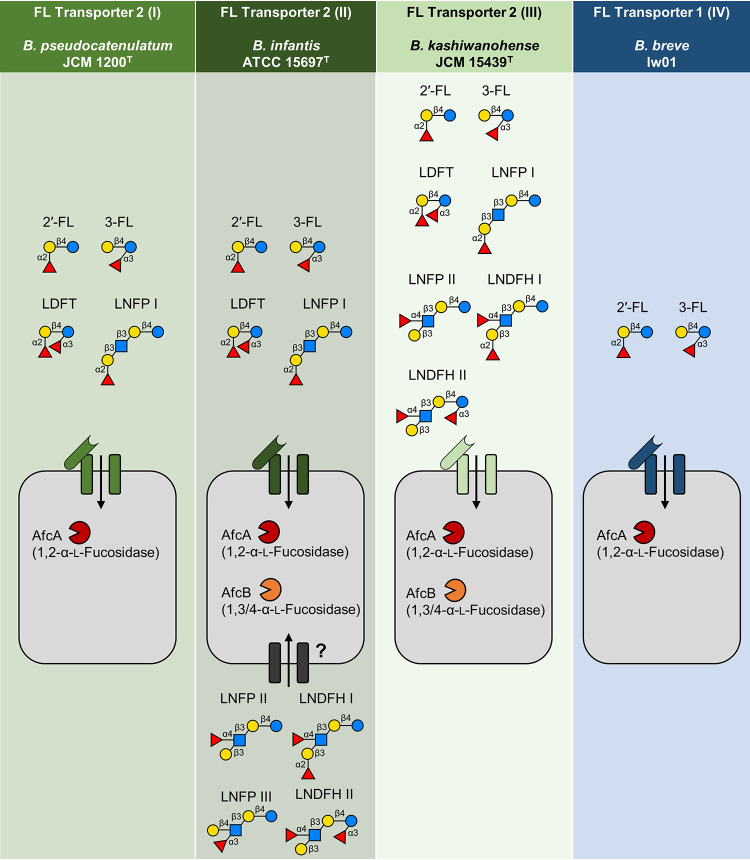
We characterized novel substrate specificities for cluster 2-III of FL transporter 2, which is present in some Bifidobacterium strains possessing AfcB homologs (Fig. 4). Previous work using pure, single sugars as a carbon source shows that this type of transporter with SBPs from cluster 2-III can import 2′-FL, 3-FL, LDFT, and LNFP I (20, 24). Interestingly, our present study revealed that the transporter from B. kashiwanohense JCM 15439T, which is associated with cluster 2-III SBPs, has an even broader specificity that includes larger, α1,3/4-fucosylated HMOs with Lewis a and b antigens, such as LNFP II, LNDFH I, and LNDFH II (Fig. 3b to d). However, this transport system was not capable of internalizing LNFP III, a Lewis x antigen-containing oligosaccharide. This is the first study to identify a transporter capable of importing LNFP II and LNDFH I/II, although a transporter that captures the degradation products of these HMOs (Lewis a trisaccharides and Lewis b tetrasaccharides) was recently identified in butyrate-producing Firmicutes (32). As shown in Fig. 4 and Fig. S5, genomic landscape analysis further revealed that all strains classified into cluster 2-III possess homologs of both AfcA and AfcB. These results suggest that the presence of intracellular fucosidases corresponds to the substrate specificity of the FL transporters. Consistent with this claim, we find that there was an apparent absence of AfcB homologs in strains possessing FL transporters with relatively limited substrate specificity (cluster 2-I, 2′-FL, 3-FL, and LDFT; cluster 1-IV, 2′-FL and 3-FL) (Fig. 4). Interestingly, these conservation patterns show strain-specific diversification, as exemplified by B. kashiwanohense, in which two strains (JCM 15439T and APCKJ1) with FL transporter cluster 2-III possess AfcB homologs but one strain (PV20-2) with FL transporter cluster 2-I does not (Fig. 4). Intracellular AfcA efficiently acts on α1,2-fucosylated HMOs like 2′-FL and LNFP I, followed by 3-FL (21, 24). Therefore, AfcA is theoretically sufficient for the metabolism of most fucosylated HMOs imported through FL transporters in clusters 2-I, 2-II, and 1-IV. However, the ability of AfcA to liberate Fuc from α1,3/4 glycosidic bonds is essentially lacking or lower than that of AfcB (21, 24, 31). Therefore, it is possible that strains with FL transporter 2-III (such as B. kashiwanohense JCM 15439T) acquired AfcA and AfcB through a gain of gene function to efficiently metabolize a diverse range of HMOs with various types of fucosylation (2′-FL, 3-FL, LDFT, LNFP I, LNFP II, LNDFH I, and LNDFH II) (Fig. 3 and 5). Conversely, AfcB is not required for strains that import only 2′-FL, 3-FL, LDFT, and LNFP I. As summarized in Fig. 6, our analyses revealed strong genotype and phenotype associations, suggesting that the diversification of FL transporters within bifidobacteria represents an adaptive strategy for HMO environments. Of note, B. longum subsp. suis BSM11-5 (cluster 2-I) possesses both AfcA and AfcB (Fig. 4). Additionally, while uptake by FL transporters in cluster 2-II is limited to 2′-FL, 3-FL, LDFT, and LNFP I (20), many strains in this cluster (mostly B. infantis) possess both AfcA and AfcB. Despite this genotype-phenotype mismatch, most B. infantis strains are capable of importing most types of HMOs, including LNFP II, LNFP III, LNDFH I, and LNDFH II (30). Future work is needed to identify the other transporters in B. infantis and B. longum subsp. suis that are responsible for importing HMOs or other sugars that require AfcB for depolymerization (Fig. 6).
FIG 6.
Intracellular fucosidase specificities are aligned with FL transporter preferences. Representative strains of each transporter type with their substrate specificities and fucosidases are shown. Strains that harbor FL transporter 2 (III) possess not only AfcA (1,2-α-l-fucosidase) but also AfcB (1,3/4-α-l-fucosidase). AfcB is generally not conserved in strains harboring transporters that can import only limited HMOs (2′-FL, 3-FL, LDFT, and LNFP I). B. infantis is an exception as it has been reported to import almost all types of HMOs through a variety of other transporters. AfcA efficiently acts on HMOs fucosylated with α-1,2 bonds (e.g., 2′-FL and LNFP I), and while it has catalytic activity on HMOs with α-1,3 bonds (e.g., 3-FL), the efficiency is low. Consequently, AfcB is predicted to be necessary to efficiently metabolize LNFP II, LNDFH I, and LNDFH II. Strains of B. infantis also possess an FL transporter 1 (IV), which is left out for clarity due to the functional overlap of both transporters.
Almost all infant gut-associated bifidobacteria can utilize LNT, a nonfucosylated HMO, while the ability to utilize fucosylated HMOs is highly strain dependent (12, 33). However, the results of our growth assays showed that B. kashiwanohense JCM 15439T did not preferentially utilize LNT and LNTri-II, an intermediate degradant of HMOs carrying LNT and LNnT as core structures (Fig. 3b and c). Instead, B. kashiwanohense JCM 15439T possesses the FL transporter that is specialized for the uptake of a broad range of fucosylated HMOs. The ability to utilize different types of fucosylated HMOs may increase fitness within the guts of infants with nonsecretor mothers (FUT2−/−). While secretor mothers produce more α1,2-fucosylated HMOs with H-antigens, such as 2′-FL and LNFP I, nonsecretor mothers produce more α1,3/4-fucosylated HMOs with Lewis antigens, such as LNFP II (4), which strains with cluster 2-III FL transporters can utilize. Despite the apparent fitness advantage conferred by the ability to consume the widest range of fucosylated HMOs, data on strains possessing cluster 2-III transporters in publicly available genomic databases are relatively limited (Fig. 1 and 4; Table S1) (20). Furthermore, the detection of B. kashiwanohense is also comparatively limited in infant data sets, especially with 16S rRNA sequencing (25, 34). However, recent studies utilizing shotgun metagenomic sequencing have shown that B. kashiwanohense is persistent and, in some cases, abundant in the infant gut microbiota (26, 35), suggesting that this transporter can provide Bifidobacterium a fitness advantage in vivo.
The results of our study, combined with previous work, highlight the importance of FL transporters for bifidobacteria in the metabolism of HMOs. Our analysis revealed that cluster (genotype) differences in FL transporters can lead to significant phenotypic variation. Furthermore, we found that the substrate specificity of each FL transporter cluster is complemented by the presence of necessary downstream fucosidase genes, suggesting a genomic adaptation to HMO-supplemented environments. By elucidating the mechanisms of fucosylated HMO uptake by bifidobacterial strains, our findings show that the diversification of the FL transporters and the matching fucosidases is a genomic signature that reflects a different adaptation strategy for HMO environments. The results of our study also highlight the strain dependency of HMO uptake phenotypes in bifidobacteria, which can be applied to the selection of effective probiotics (Bifidobacterium) and prebiotics (HMOs) for individualized clinical interventions and formula milk.
MATERIALS AND METHODS
Chemicals.
Fuc, Glc, Gal, 2-AA (2-anthranilic acid), and sodium cyanoborohydride were purchased from Nacalai Tesque (Kyoto, Japan). GlcNAc and Lac were purchased from Fujifilm Wako Pure Chemical (Osaka, Japan). The following sugars were purchased from Dextra Laboratory (Reading, UK): LNnT and LNFP I. 2′-FL, 3-FL, LDFT, and LNT were provided as gifts from Glycom A/S (Hørsholm, Denmark). LNTri-II was obtained through the digestion of LNnT using Bga2A, a β-1,4-galactosidase from B. infantis (36). LNT and LNTri-II were further purified by size exclusion chromatography (Toyopearl HW-40C, 20 mm by 600 mm; Tosoh Corporation, Tokyo, Japan). LNFP II, LNFP III, LNDFH I, and LNDFH II were purchased from Carbosynth (Berkshire, UK), and isomaltoheptaose was purchased from Seikagaku Kogyo (Tokyo, Japan). All other reagents that we used were of analytical grade.
Preparation of oligosaccharides from human milk.
Milk samples were collected from healthy Japanese mothers who had not taken any antibiotics for at least 1 month before collection at Nagao Midwife Clinics (Kyoto, Japan). Informed consent was obtained from all mothers. The study was reviewed and approved by the Ethics Committee of Kyoto University (R0046-1) and the University of Shiga Prefecture (71-3) and was performed according to the Declaration of Helsinki. HMOs were purified from the collected human milk samples as described previously (30). During the purification process, sialyl oligosaccharides were eluted near the void fractions as polymeric compounds and therefore were absent from the HMO mixture used in this study. Lac (approximately 6% of the mixture by molar ratio), a contaminant derived from the HMO purification process, was also detected.
Bacterial strains and culture conditions in HMO-supplemented medium.
B. kashiwanohense JCM 15439T and B. pseudocatenulatum JCM 1200T were obtained from the Japan Collection of Microorganisms (RIKEN BioResource Research Center, Tsukuba, Japan). The MS554 strain is a previously constructed B. longum JCM 31944 ΔlnbX ΔgltA double mutant heterologously expressing genes that encode intracellular exo-glycosidases for HMO depolymerization (1,2-α-l-fucosidase gene [Blon_2335], N-acetylglucosaminidase [Blon_0459], LNT β-1,3-galactosidase [Blon_2106], 1,3/4-α-l-fucosidase [Blon_2336], and sialidase gene [Blon_2348] from B. infantis) (20). The heterologous expression strain was previously constructed by introducing a plasmid carrying the FL transporter 2 homolog genes (BBKW_1838–1840) from B. kashiwanohense JCM 15439T into MS554, and as a negative control, an empty vector was introduced into the MS554 strain previously (20). All bifidobacterial strains were routinely cultured in Gifu anaerobic medium (GAM) broth (Nissui Pharmaceutical, Tokyo, Japan) at 37°C under anaerobic conditions using the AnaeroPack system (Mitsubishi Gas Chemical Co., Tokyo, Japan). Culturing experiments to examine carbohydrate utilization by bifidobacteria were performed using de Man-Rogosa-Sharpe medium containing 0.02% (wt/vol) cysteine-HCl and 0.34% (wt/vol) sodium ascorbate (MRS-CS). As a sole carbon source, 1% (wt/vol) HMOs were used instead of glucose. Each strain was inoculated into the medium with an initial optical density at 600 nm (OD600) of 0.02. All cultures were incubated under anaerobic conditions at 37°C for 24 h. Growth was monitored by measuring the OD600 at each time point. All culturing experiments were performed in biological triplicate.
Sugar concentration analysis.
Culture supernatants were collected by centrifuging the bacterial incubation samples at each time point and stored at −30°C until use. HPLC analysis was performed as described previously (30, 37), in which the samples were thawed and mixed with isomaltoheptaose (internal standard), the sugars were fluorescence labeled with 2-AA, and the reaction mixtures were desalted by solid-phase extraction (30, 38).
Identification of oligosaccharides using enzymatic treatments.
Enzymatic assays were used to identify LNTri-II and to differentiate between LNFP II and LNFP III, which are eluted as a combined peak during HPLC analysis. During HPLC analysis, the fraction containing LNFP II and LNFP III (retention time of around 55 to 57 min) and the fraction containing LNTri-II (retention time of 35 to 37 min) were manually collected. Acetonitrile was evaporated under a constant nitrogen gas stream and then heated at 40°C for 15 min, and the samples were freeze-dried overnight and dissolved in water. To differentiate between LNFP II and LNFP III, 1,3/4-α-l-fucosidase (AfcB), purified as described previously (29), was used. To identify LNTri-II, β-1,3-N-acetylglucosaminidase I (BbhI), purified as described previously (39), was used. Each 100-μL enzyme reaction mixture contained the following: 50 μL of the fractionated sample, either AfcB or BbhI (final concentration of 100 μg/mL), and sodium acetate (final concentration of 50 mM [pH 6.0]). The mixture was incubated for 21 h at 30°C, followed by 5 min at 95°C to halt the reaction. The resulting mixture was analyzed by HPLC as described above.
Analysis of fucosidase activity using cell extracts.
B. kashiwanohense JCM 15439T and B. pseudocatenulatum JCM 1200T were cultured in MRS-CS medium containing 0.5% 2′-FL until the OD600 reached 0.5 to 0.7, after which the bacterial cells were harvested, washed twice with 50 mM sodium phosphate buffer (pH 6.5) containing 1 mM 2-mercaptoethanol, disrupted by sonication, and clarified by centrifugation. For fucosyllactose degradation ability, the cell extracts (protein concentration of 64.1 μg/mL) were incubated at 37°C with 3 mM 2′-FL, 3-FL, and LNFP II in the same buffer, and the samples were then analyzed by thin-layer chromatography in a solution containing 1-butanol, acetic acid, and water (2:1:1 ratio) as the developing solvent, as described previously (31, 40).
Genomic conservation and phylogenetic analysis of FL2-BP, AfcA, and AfcB homologs.
The conservation of FL2-BP homologs was analyzed with tBLASTn using the NCBI genomic database available on 25 January 2021 (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The analysis also included the B. longum SC596 genome (22). The search was performed using the amino acid sequence of the FL2-BP protein of B. infantis ATCC 15697T (Blon_2202) against the available draft and complete genome sequences of bifidobacterial strains, and sequences of proteins that share ≥60% amino acid identity were used for phylogenetic analysis. For homologs assigned to the B. longum group, only the search results for which the subspecies were shown were used for analysis. The search retrieved 89 applicable homologs from 69 bifidobacterial strains. The phylogenetic tree of FL2-BP homologs was created using RAxML v8.1.20 (41) with 1,000 bootstrap iterations following sequence alignment in Clustal W with default settings (www.genome.jp/tools-bin/clustalw). For the strains possessing FL2-BP homologs, the presence of α-1,2-l-fucosidase (AfcA [GH95] [Blon_2335]) and α-1,3/4-l-fucosidase (AfcB [GH29] [Blon_2336]) was also analyzed as described above.
Data availability.
The data supporting the conclusions of this study are available and included within the article.
ACKNOWLEDGMENTS
We thank Saeko Nagao (Nagao Midwife Clinic) for her help with collecting breast milk samples and Glycom A/S for providing LNT, 2′-FL, 3-FL, and LDFT.
This study was supported in part by grants-in-aid for JSPS Research Fellows (19J14598 to Miriam N. Ojima), JSPS-KAKENHI (21K14770 to Mikiyasu Sakanaka and 19K22277 and 21H02116 to Takane Katayama), and the Mishima Kaiun Memorial Foundation (to Motomitsu Kitaoka and Takane Katayama).
Mikiyasu Sakanaka and Takane Katayama conceived and designed the study. Miriam N. Ojima, Mikiyasu Sakanaka, and Maher Abou Hachem contributed to the bioinformatics analyses. Junko Hirose collected the breast milk samples, and Miriam N. Ojima, Mikiyasu Sakanaka, Takane Katayama, and Motomitsu Kitaoka were responsible for purifying the HMOs from the samples. Miriam N. Ojima, Yuya Asao, Mikiyasu Sakanaka, Aina Gotoh, Tadasu Urashima, Satoru Fukiya, and Atsushi Yokota contributed to the culturing experiments and sugar concentration analysis using different Bifidobacterium strains. Yuya Asao and Toshihiko Katoh purified BbhI and AfcB and performed enzymatic digestion. Aruto Nakajima performed the cell-free assays. Miriam N. Ojima, Yuya Asao, and Mikiyasu Sakanaka contributed to the draft, and Miriam N. Ojima wrote the manuscript. Maher Abou Hachem, Mikiyasu Sakanaka, and Takane Katayama supervised the study and edited the manuscript. All authors contributed to manuscript revision and approved the submitted version.
Employment of Mikiyasu Sakanaka at Kyoto University is supported by Morinaga Milk Industry Co. Ltd. We declare no other competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Mikiyasu Sakanaka, Email: sakanaka.mikiyasu.5e@kyoto-u.ac.jp.
Hideaki Nojiri, University of Tokyo.
REFERENCES
- 1.Urashima T, Asakuma S, Leo F, Fukuda K, Messer M, Oftedal OT. 2012. The predominance of type I oligosaccharides is a feature specific to human breast milk. Adv Nutr 3:473S–482S. 10.3945/an.111.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr 20:699–722. 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 3.Bode L. 2012. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22:1147–1162. 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGuire MK, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, Prentice AM, Kvist LJ, Otoo GE, Brooker SL, Price WJ, Shafii B, Placek C, Lackey KA, Robertson B, Manzano S, Ruíz L, Rodríguez JM, Pareja RG, Bode L. 2017. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr 105:1086–1100. 10.3945/ajcn.116.139980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. 2000. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr 71:1589–1596. 10.1093/ajcn/71.6.1589. [DOI] [PubMed] [Google Scholar]
- 6.Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, Ogawa E, Kodama H, Yamamoto K, Yamada T, Matsumoto S, Kurokawa K. 2016. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun 7:11939. 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrido D, Dallas DC, Mills DA. 2013. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology (Reading) 159:649–664. 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcobal A, Sonnenburg JL. 2012. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect 18:12–15. 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tannock GW, Lawley B, Munro K, Pathmanathan SG, Zhou SJ, Makrides M, Gibson RA, Sullivan T, Prosser CG, Lowry D, Hodgkinson AJ. 2013. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl Environ Microbiol 79:3040–3048. 10.1128/AEM.03910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh T, Ojima MN, Sakanaka M, Ashida H, Gotoh A, Katayama T. 2020. Enzymatic adaptation of Bifidobacterium bifidum to host glycans, viewed from glycoside hydrolyases and carbohydrate-binding modules. Microorganisms 8:481. 10.3390/microorganisms8040481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katayama T. 2016. Host-derived glycans serve as selected nutrients for the gut microbe: human milk oligosaccharides and bifidobacteria. Biosci Biotechnol Biochem 80:621–632. 10.1080/09168451.2015.1132153. [DOI] [PubMed] [Google Scholar]
- 12.Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao J-Z, Kitaoka M, Katayama T. 2020. Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients 12:71. 10.3390/nu12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez CI, Martiny JBH. 2020. Evolutionary relationships among bifidobacteria and their hosts and environments. BMC Genomics 21:26. 10.1186/s12864-019-6435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C, van Sinderen D, Ventura M. 2016. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol 82:980–991. 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. 2011. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10:507–514. 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostopoulos I, Elzinga J, Ottman N, Klievink JT, Blijenberg B, Aalvink S, Boeren S, Mank M, Knol J, de Vos WM, Belzer C. 2020. Akkermansia muciniphila uses human milk oligosaccharides to thrive in the early life conditions in vitro. Sci Rep 10:14330. 10.1038/s41598-020-71113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama T, Sakuma A, Kimura T, Makimura Y, Hiratake J, Sakata K, Yamanoi T, Kumagai H, Yamamoto K. 2004. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-α-l-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95). J Bacteriol 186:4885–4893. 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pokusaeva K, Fitzgerald GF, van Sinderen D. 2011. Carbohydrate metabolism in bifidobacteria. Genes Nutr 6:285–306. 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrido D, Kim JH, German JB, Raybould HE, Mills DA. 2011. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS One 6:e17315. 10.1371/journal.pone.0017315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakanaka M, Hansen ME, Gotoh A, Katoh T, Yoshida K, Odamaki T, Yachi H, Sugiyama Y, Kurihara S, Hirose J, Urashima T, Xiao JZ, Kitaoka M, Fukiya S, Yokota A, Lo Leggio L, Abou Hachem M, Katayama T. 2019. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci Adv 5:eaaw7696. 10.1126/sciadv.aaw7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sela DA, Garrido D, Lerno L, Wu S, Tan K, Eom HJ, Joachimiak A, Lebrilla CB, Mills DA. 2012. Bifidobacterium longum subsp. infantis ATCC 15697 α-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol 78:795–803. 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrido D, Ruiz-Moyano S, Kirmiz N, Davis JC, Totten SM, Lemay DG, Ugalde JA, German JB, Lebrilla CB, Mills DA. 2016. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci Rep 6:35045. 10.1038/srep35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA 105:18964–18969. 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James K, Bottacini F, Contreras JIS, Vigoureux M, Egan M, O’Connell Motherway M, Holmes E, van Sinderen D. 2019. Metabolism of the predominant human milk oligosaccharide fucosyllactose by an infant gut commensal. Sci Rep 9:15427. 10.1038/s41598-019-51901-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita H, Nakano A, Onoda H, Toh H, Oshima K, Takami H, Murakami M, Fukuda S, Takizawa T, Kuwahara T, Ohno H, Tanabe S, Hattori M. 2011. Bifidobacterium kashiwanohense sp. nov., isolated from healthy infant faeces. Int J Syst Evol Microbiol 61:2610–2615. 10.1099/ijs.0.024521-0. [DOI] [PubMed] [Google Scholar]
- 26.Busi SB, de Niels L, Habier J, Wampach L, Fritz JV, Heintz-Buschart A, May P, Halder R, de Beaufort C, Wilmes P. 2021. Persistence of birth mode-dependent effects on gut microbiome composition, immune system stimulation and antimicrobial resistance during the first year of life. ISME Commun 1:8. 10.1038/s43705-021-00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunesova V, Lacroix C, Schwab C. 2016. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol 16:248. 10.1186/s12866-016-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrido D, Ruiz-Moyano S, Mills DA. 2012. Release and utilization of N-acetyl-D-glucosamine from human milk oligosaccharides by Bifidobacterium longum subsp. infantis. Anaerobe 18:430–435. 10.1016/j.anaerobe.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. 2009. Two distinct α-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19:1010–1017. 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 30.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, Kumagai H, Ashida H, Hirose J, Kitaoka M. 2011. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem 286:34583–34592. 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakurama H, Fushinobu S, Hidaka M, Yoshida E, Honda Y, Ashida H, Kitaoka M, Kumagai H, Yamamoto K, Katayama T. 2012. 1,3-1,4-α-l-Fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains. J Biol Chem 287:16709–16719. 10.1074/jbc.M111.333781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichler MJ, Yamada C, Shuoker B, Alvarez-Silva C, Gotoh A, Leth ML, Schoof E, Katoh T, Sakanaka M, Katayama T, Jin C, Karlsson NG, Arumugam M, Fushinobu S, Abou Hachem M. 2020. Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways. Nat Commun 11:3285. 10.1038/s41467-020-17075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomson P, Medina DA, Garrido D. 2018. Human milk oligosaccharides and infant gut bifidobacteria: molecular strategies for their utilization. Food Microbiol 75:37–46. 10.1016/j.fm.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez-Gutierrez P, Lacroix C, Jaeggi T, Zeder C, Zimmerman MB, Chassard C. 2015. Bifidobacteria strains isolated from stools of iron deficient infants can efficiently sequester iron. BMC Microbiol 15:3. 10.1186/s12866-014-0334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan W, Luo B, Zhang X, Ni Y, Tian F. 2021. Association and occurrence of bifidobacterial phylotypes between breast milk and fecal microbiomes in mother-infant dyads during the first 2 years of life. Front Microbiol 12:669442. 10.3389/fmicb.2021.669442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida E, Sakurama H, Kiyohara M, Nakajima M, Kitaoka M, Ashida H, Hirose J, Katayama T, Yamamoto K, Kumagai H. 2012. Bifidobacterium longum subsp. infantis uses two different β-galactosidases for selectively degrading type-1 and type-2 human milk oligosaccharides. Glycobiology 22:361–368. 10.1093/glycob/cwr116. [DOI] [PubMed] [Google Scholar]
- 37.Gotoh A, Katoh T, Sakanaka M, Ling Y, Yamada C, Asakuma S, Urashima T, Tomabechi Y, Katayama-Ikegami A, Kurihara S, Yamamoto K, Harata G, He F, Hirose J, Kitaoka M, Okuda S, Katayama T. 2018. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci Rep 8:13958. 10.1038/s41598-018-32080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anumula KR. 2006. Advances in fluorescence derivatization methods for high-performance liquid chromatographic analysis of glycoprotein carbohydrates. Anal Biochem 350:1–23. 10.1016/j.ab.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 39.Miwa M, Horimoto T, Kiyohara M, Katayama T, Kitaoka M, Ashida H, Yamamoto K. 2010. Cooperation of β-galactosidase and β-N-acetylhexosaminidase from bifidobacteria in assimilation of human milk oligosaccharides with type 2 structure. Glycobiology 20:1402–1409. 10.1093/glycob/cwq101. [DOI] [PubMed] [Google Scholar]
- 40.Sakurama H, Kiyohara M, Wada J, Honda Y, Yamaguchi M, Fukiya S, Yokota A, Ashida H, Kumagai H, Kitaoka M, Yamamoto K, Katayama T. 2013. Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J Biol Chem 288:25194–25206. 10.1074/jbc.M113.484733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5. Download AEM.01437-21-s0001.pdf, PDF file, 0.4 MB (372.3KB, pdf)
Table S1. Download AEM.01437-21-s0002.xlsx, XLSX file, 0.01 MB (13.5KB, xlsx)
Data Availability Statement
The data supporting the conclusions of this study are available and included within the article.