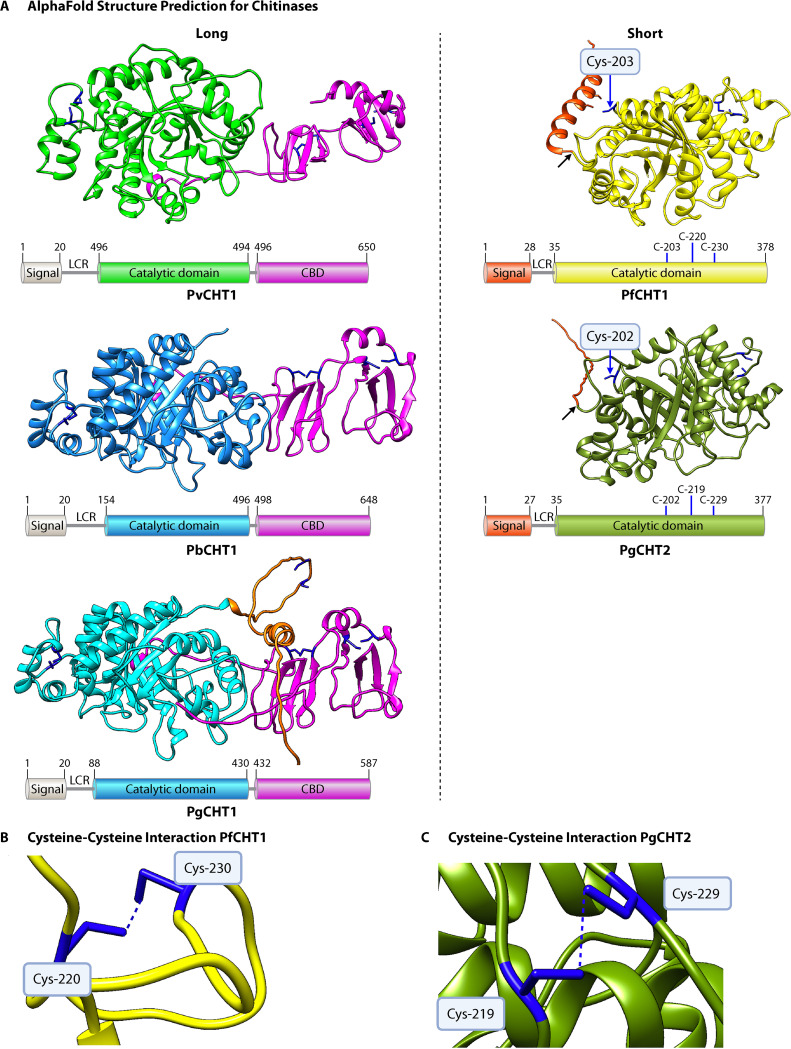
FIG 4.
AlphaFold predictions for the structures of chitinases in P. vivax, P. berghei, P. gallinaceum, and P. falciparum. (A) Chitinases are categorized as either long or short, with the long chitinases having a chitin-binding domain. The long chitinases were trimmed to exclude the signal peptide and low-complexity regions, except that some of the low-complexity region is still included for PgCHT1. A schematic representation of long chitinases is included under the structures, showing a distinction between catalytic and chitin-binding domains. The signal peptide sequences for all the represented protein structures were predicted using SignalP 5.0 (69). For short chitinases, the three cysteines within the catalytic domain are included in the schematic, and the free, surface-exposed cysteine is labeled and marked with a blue arrow on the model. The signal peptide cleavage site is marked with a black arrow for the short chitinases. The low-complexity region is indicated on the schematics with the abbreviation LCR. (B) Closer view of the projected disulfide bridge formed between cysteines at amino acid positions 220 and 230 in PfCHT1. (C) Closer view of the projected disulfide bridge formed between cysteines at amino acid positions 219 and 229 in PgCHT2.