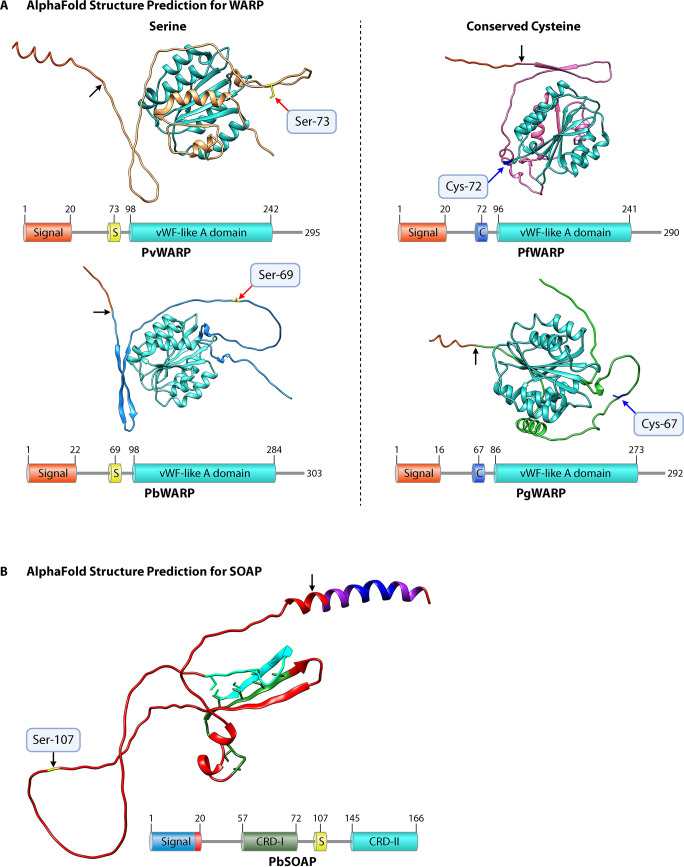
FIG 5.
AlphaFold predictions for the structures of WARP and SOAP in P. vivax, P. berghei, P. gallinaceum, and P. falciparum. (A) Both P. vivax and P. berghei have a serine, shown in yellow, rather than a conserved cysteine, marked with a red arrow and labeled. Both P. falciparum and P. gallinaceum have a conserved cysteine, marked with a blue arrow and labeled. The signal peptide cleavage site is marked with a black arrow; the signal peptide has been trimmed in all proteins. The associated protein schematic shows functional components of protein. A conserved von Willebrand factor (vWF)-like A domain is seen across all four species, predicted using PROSITE (70), shown in turquoise. (B) "Unstructured" PbSOAP is representative of the structure prediction for SOAP for all species. The signal peptide cleavage site is marked with black arrow, and serine is shown in yellow and is present in all Plasmodium species with long chitinases (except glutamine in P. malariae). Except for the signal peptide, the structure prediction for SOAP was unconfident. The schematic shows the presence of two cysteine-rich domains (I and II) having six cysteine residues in each domain. These domains are conserved in almost all the Plasmodium species except for one cysteine residue present in between the region of the two domains in the members of only the Laverania subgenus and avian-infecting species.