Figure 1:
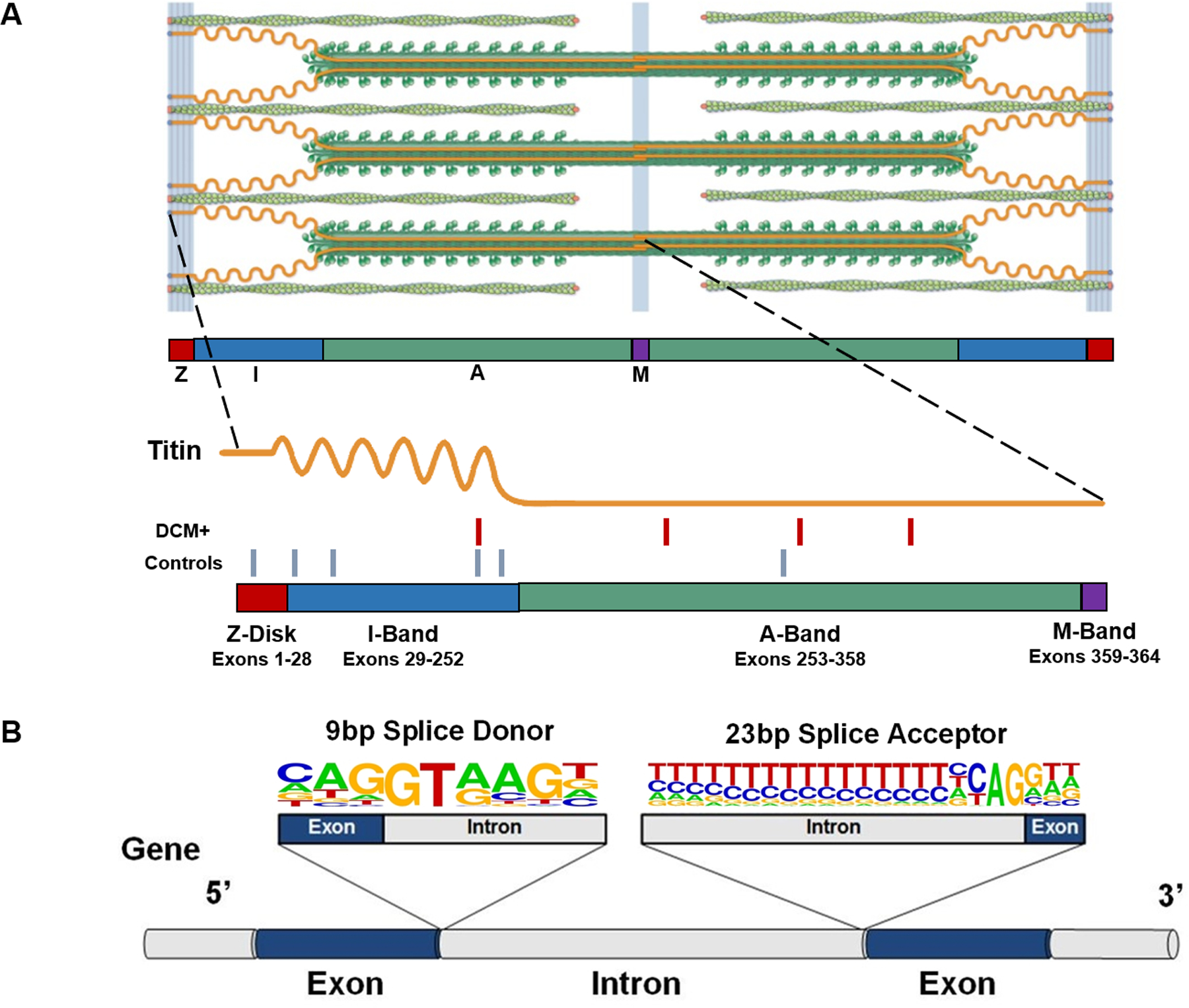
(A) The giant protein titin is a key component of the cardiac sarcomere. A standard cardiac sarcomere is composed of thick filaments (dark green), thin filaments (light green), and a filament composed of the giant protein titin (orange). A single full-length titin molecule spans the Z-disk, I-band, A-band and M-band regions, serving as a spring that preserves the precise structural integrity of the sarcomere. Being the largest human protein, the TTN gene contains 364 exons, corresponding with 726 exon-intron junctions, each of which need to be correctly recognized and processed to create a functional full-length polypeptide. The sarcomere image is adapted from Herman et al.7 with permission. The location of 4 non-canonical splice-altering variants identified in 203 DCM cases (red hash marks) and 6 non-canonical splice altering variants in 3329 controls (blue hash marks) is indicated on the schematic. (B) Splice donor and splice acceptor sites are defined by conserved 9 bp and 23 bp sequences at exon-intron junctions. A gene segment containing two exons (blue) and flanking introns (grey) with consensus 9 bp splice donor and 23 bp splice acceptor sequences is shown. The nucleotide letter sizes denote usage of that base in splice sites across the genome. Dinucleotides GT (donor site) and AG (acceptor site) are invariant, and variants that disrupt these canonical splicing residues are readily classified as TTNtv. Variants located in the other positions of these sequences may also affect splicing, but are often left labeled as VUS. TTN = Titin; bp = base pair; TTNtv = titin truncating variant; VUS = variants of unknown significance.
