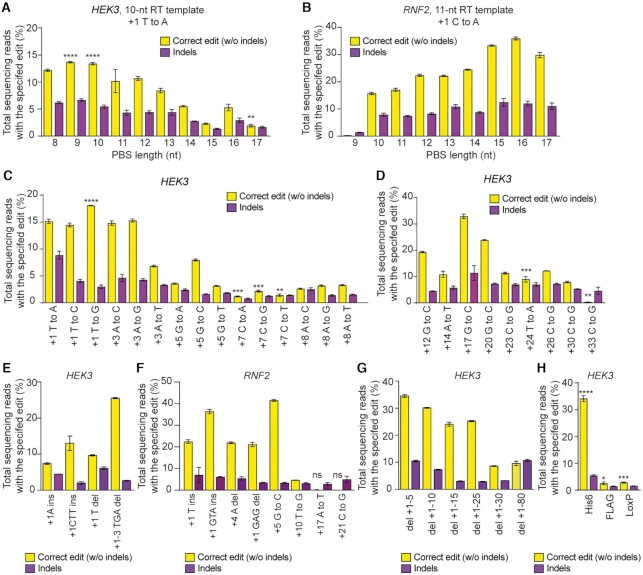
Figure 2.
Prime editing of genomic DNA in H9-iPE2 cells by PE3. (A, B) PE3 editing efficiencies at HEK3 and RNF2 genomic sites with pegRNAs containing varying PBS lengths. (C) Efficiencies of all 12 types of transition and transversion edits at the indicated positions in the HEK3 site. (D) Efficiencies of long distance edits at the HEK3 site using a 34-nt RT template. (E, F) Editing efficiencies for the generation of intended indels at the HEK3 and RNF2 genomic sites. Yellow, desired indel; purple, undesired indel. (G) Editing efficiencies for the generation of targeted deletions of 5–80 bp at the HEK3 site. (H) Editing efficiencies for the generation of targeted insertions of a His6 tag, Flag epitope tag, and loxP site at the HEK3 site. The editing efficiency is indicated as the percentage of total sequencing reads that contain the intended edit and do not contain unintended indels. Mean ± s.d. of n = 3 independent biological replicates.