Figure 1.
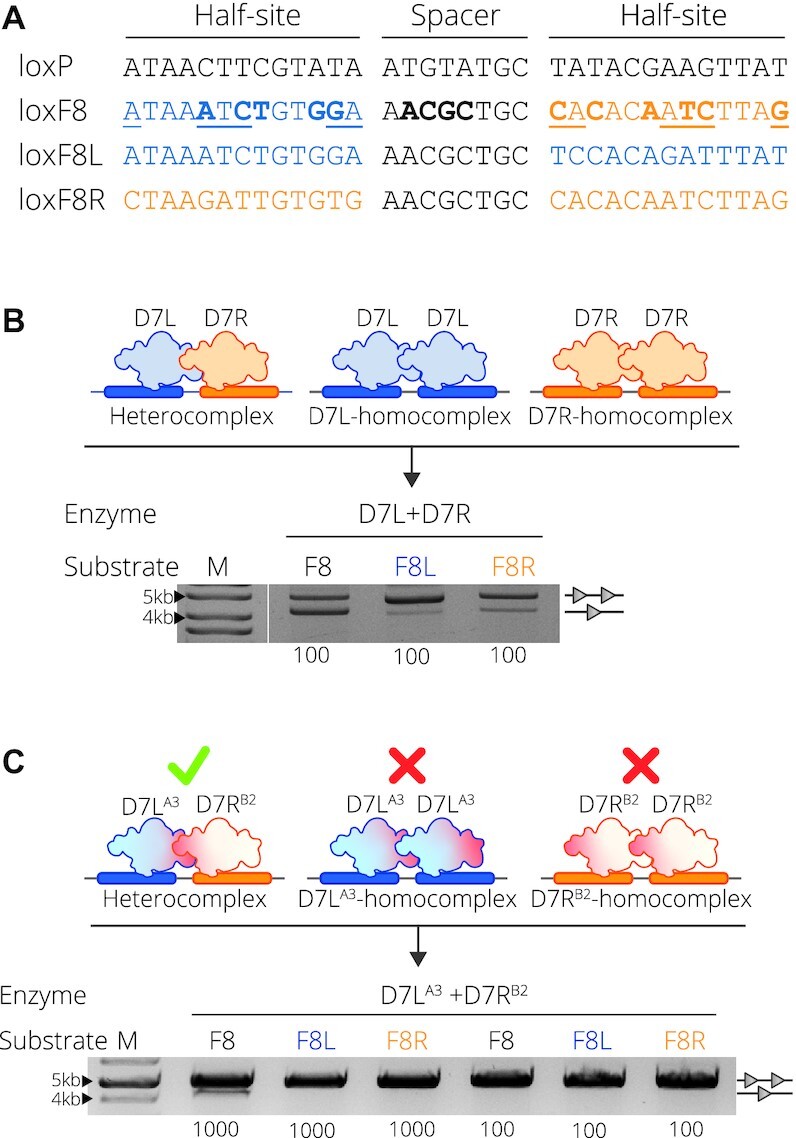
D7 target site substrates and recombination activity. (A) Full target sequences of loxP, loxF8, loxF8L (blue) and loxF8R (orange) separated into the two half-sites flanking the spacer sequence. The asymmetric loxF8 site is displayed with the left half-site as blue and the right half site as orange correlating to the full symmetric sites loxF8L and loxF8R, respectively. Underlined nucleotides in the sequence represent asymmetric positions and the bold nucleotides represent positions differing from the loxP sequence. (B) Complex formation and resulting activity of the D7L and D7R monomers. Upper panel: D7L (blue recombinase) and D7R (orange recombinase) heterocomplex formation with the loxF8 asymmetric site (blue and orange rectangles) and the homocomplex of D7L and D7R with the symmetric sites of loxF8L (blue rectangles) and loxF8R (orange rectangles), respectively. Lower panel: Bacterial assay of recombination activity of co-expressed D7L and D7R (Enzyme) on target sites loxF8, loxF8L and loxF8R (indicated with Substrate F8, F8L and F8R, respectively) all induced with a concentration of 100 μg/ml l-Arabinose (listed along the bottom of the gel image). Recombination is signified by the band aligned with the single triangle and non-recombined events are indicated by two triangles. M = 10 kb ladder (GeneRuler DNA Ladder Mix 10 kb). (C) Upper panel: Schematic of the desired recombination events when mutations (indicated in red) are applied to the molecules. Mutations from the group A3 are applied to D7L and B2 is applied to D7R indicated by the superscript (20). Desired recombination is depicted as such: when both mutated monomers are present, they are able to form an active heterocomplex (green check) with the asymmetric loxF8 site (blue and orange rectangles); in isolation they cannot form functional homotetrameric complexes (red X) with symmetric sites. Lower panel: recombination activity of D7LA3 and D7RB2 on the three substrates. Concentration (μg/ml) of L-Arabinose used for induction is indicated along the bottom. M = 10 kb ladder (GeneRuler DNA Ladder Mix 10kb).
