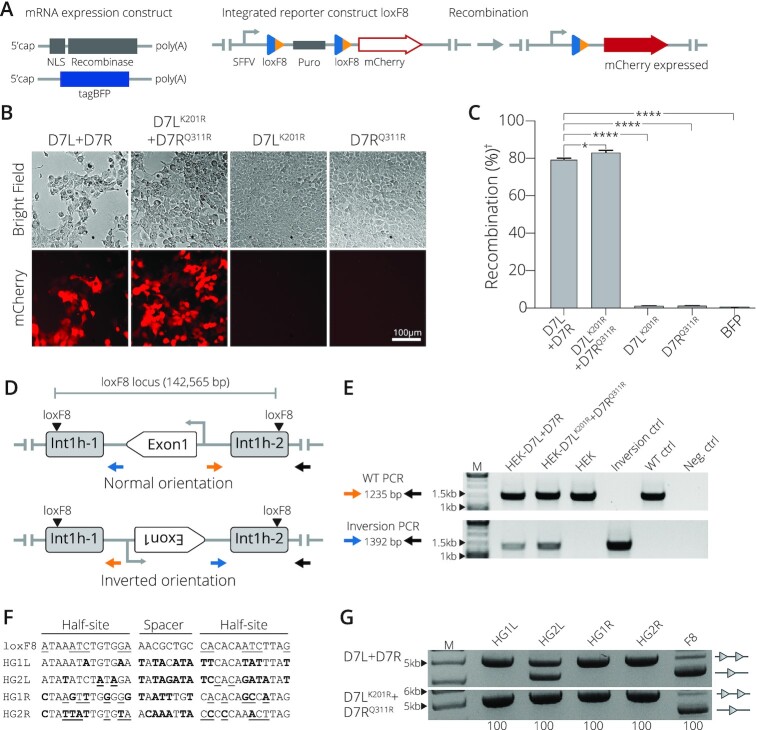
Figure 3.
Obligate D7LK201R+ D7RQ311R activity in mammalian cells. (A) Schematic presentation of mRNA expression constructs and the reporter HEK293T cell line. Employed mRNAs with indicated features (5′cap and polyA tail) expressing a nuclear localization signal (NLS) fused to the recombinase and the tagBFP mRNA are shown. The stable reporter cell line harbors two loxF8 sites (blue/orange triangles) that flank a puromycin selection gene (puro). Once successfully excised by recombination, mCherry is expressed from the SFFV promoter (arrow). (B) Recombination assays in mammalian cells. Representative brightfield and mCherry fluorescent images of reporter HEK293T cells transfected with indicated recombinases. The 100 μm scale bar is indicated. (C) Quantification of the recombination efficiencies 48 h after transfection of HEK293T reporter cells with indicated recombinases, analyzed by FACS (Error bar represents the SD of experiments performed in biological triplicates, n = 3). Recombination efficiency determined by percent of cells expressing mCherry of the total of BFP positive cells. (†) indicates normalization to BFP signal. (*) and (****) indicate P < 0.05 and P < 0.0001, respectively. (D) Scheme of the genomic inversion detection PCR. The WT orientation is indicated by the amplicon produced by primers colored in blue and gray. The inverted orientation is indicated by primers in orange and gray. Exon 1 of the factor VIII gene and the genomic loxF8 target sites are indicated. (E) Gel showing PCR results with primers detecting the WT orientation (top panel) and primers detecting the inverted orientation (bottom panel). Results for transfections with indicated mRNAs are shown. Controls included HEK293T cells that were transfected with BFP mRNA only (non-treated), int1h patient DNA carrying the inversion of exon1 (Inverted-Ctrl), non-treated cells (WT-Ctrl) and water only (H2O). M = GeneRuler DNA Ladder Mix 10 kb (F) Sequences of predicted off-target sites in the human genome with high sequence similarity to loxF8L (HG1L and HG2L) and loxF8R (HG1R and HG2R). Bold nucleotides indicate nucleotides differing from the loxF8 target sequence and underlined nucleotides indicate asymmetry. (G) Bacterial assay for recombination activity of D7L + D7R compared to the activity of D7LK201R+ D7RQ311R on the indicated human off-target sites. Recombination activity on loxF8 is shown as control. Bands of non-recombined plasmids are indicated by a line with two triangles, while recombined bands are marked by a line with one triangle. 100 (μg/ml) of l-Arabinose used for recombinase induction is indicated along the bottom. M = GeneRuler DNA Ladder Mix 10 kb.