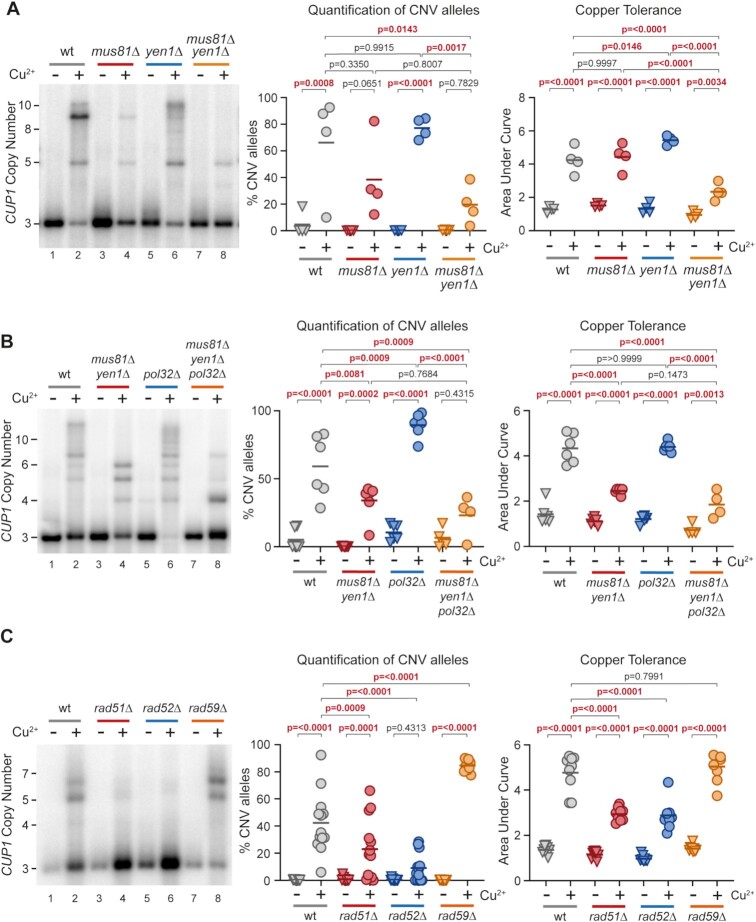
Figure 5.
Importance of SSEs and Rad proteins in CUP1 CNV. (A) Southern analysis of CUP1 copy number in 3xCUP1 wild-type (wt), mus81Δ, yen1Δ and mus81Δ yen1Δ cells after 10 generations ±0.3 mM CuSO4. Quantification shows the percentage of alleles deviating from the parental copy number, P-values calculated by one-way ANOVA. Copper adaptation was assessed by treated cells with varying concentrations of CuSO4 and incubating for 3 days. Final OD600 was plotted against [CuSO4] and copper tolerance quantified as area-under-curve for each culture; P-values were calculated by one-way ANOVA; n = 4. (B) Southern blot analysis of CUP1 copy number and adaptation test in 3xCUP1 wild-type (wt), mus81Δ yen1Δ, pol32Δ and mus81Δ yen1Δ pol32Δ cells after 10 generations ±0.3 mM CuSO4, analysed as in (A), n = 6. (C) Southern blot analysis of CUP1 copy number and adaptation test in 3xCUP1 wild-type (wt), rad51Δ, rad52Δ and rad59Δ cells after 10 generations ±0.3 mM CuSO4, analysed as in (A), n = 12.