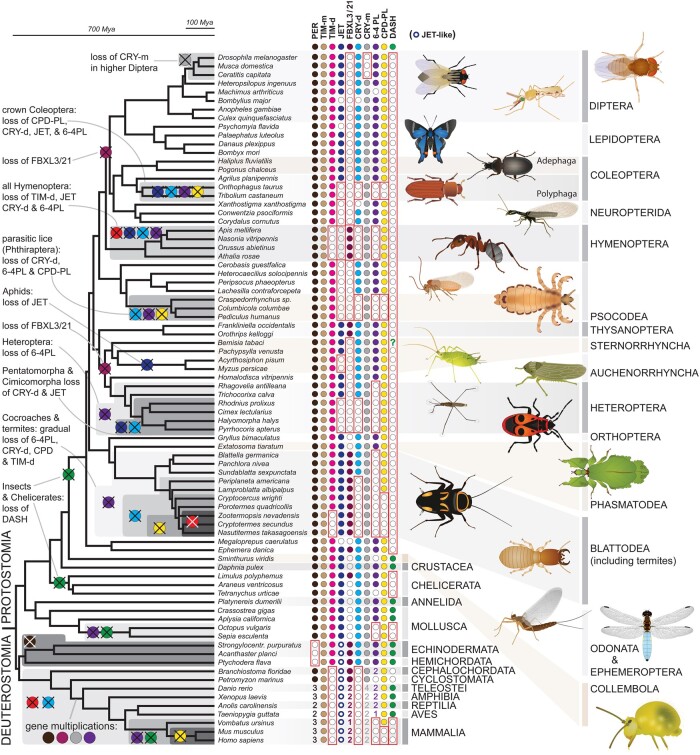
Fig. 2.
Circadian clock gene losses mapped on the bilaterian phylogeny. Representative insect species are shown at the terminal nodes with indicated gene presence (full circle) or absence (empty circle) where the lineage-specific losses are highlighted with red rectangles (see details in supplementary tables 1 and 2, Supplementary Material online). The phylogenetic tree corresponds to a consensus of recent phylogenomic studies (Misof et al. 2014; Johnson et al. 2018; Kawahara et al. 2019; McKenna et al. 2019; Wipfler et al. 2019). Filled circles indicate the presence of PER (black), TIM-m (brown), TIM-d (bright magenta), JET (dark blue), FBXL3/21 (light purple), CRY-m (grey), CRY-d (blue), 6-4 PL (deep purple), CPD-PL (yellow), and DASH-type cryptochrome (green). For phylogenetic relationship see supplementary figures 1–4, Supplementary Material online. Numbers indicate the presence of multiple paralogs in one taxon. The question mark indicates a suspicious occurrence of DASH in Bemisia. Supplementary figure 16, Supplementary Material online, illustrates clear relatedness of this sequence with DASH from plants and fungi which can either be explained as contamination or as a horizontal gene transfer (HGT) from plant to insect. The latter would be consistent with a recent HGT of a plant detoxification component to Bemisia (Xia et al. 2021).