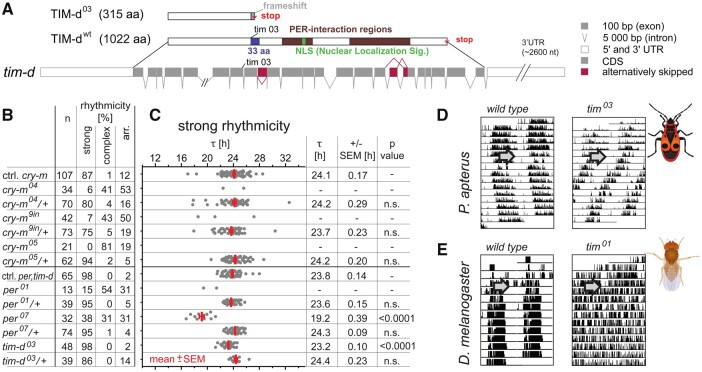
Fig. 4.
Either cry-m or per depletion completely abolishes circadian rhythmicity in P. apterus, whereas tim-d mutants demonstrate robust rhythmicity with significantly shorter τ. (A) Schematic representation of tim-d gene structure with coding regions, alternative splicing, and engineered mutation (tim03). Corresponding wt and mutant proteins are shown with major functional domains highlighted (for details, see supplementary figs. 13 and 14, Supplementary Material online). Alternative splicing of tim-d was detected in exons 9, 17, and 18. (B) Summary indicating the number and rhythmicity of the tested mutant and heterozygous animals compared with corresponding control siblings. (C) Individual τ values are plotted as a dot for each male; red bars depict means ± SEMs (calculated only if >10% individuals were rhythmic). Statistical difference from the controls is shown as P-value (Kruskal–Wallis test with Dunn’s post hoc). Double-plotted actogram of (D) wt and tim03 P. apterus compared with (E) D. melanogaster wt (Canton S) and tim01 mutant (arrow indicates the beginning of constant darkness).