Abstract
The mutualism between the giant tubeworm Riftia pachyptila and its endosymbiont Candidatus Endoriftia persephone has been extensively researched over the past 40 years. However, the lack of the host whole-genome information has impeded the full comprehension of the genotype/phenotype interface in Riftia. Here, we described the high-quality draft genome of Riftia, its complete mitogenome, and tissue-specific transcriptomic data. The Riftia genome presents signs of reductive evolution, with gene family contractions exceeding expansions. Expanded gene families are related to sulfur metabolism, detoxification, antioxidative stress, oxygen transport, immune system, and lysosomal digestion, reflecting evolutionary adaptations to the vent environment and endosymbiosis. Despite the derived body plan, the developmental gene repertoire in the gutless tubeworm is extremely conserved with the presence of a near intact and complete Hox cluster. Gene expression analyses establish that the trophosome is a multifunctional organ marked by intracellular digestion of endosymbionts, storage of excretory products, and hematopoietic functions. Overall, the plume and gonad tissues both in contact to the environment harbor highly expressed genes involved with cell cycle, programed cell death, and immunity indicating a high cell turnover and defense mechanisms against pathogens. We posit that the innate immune system plays a more prominent role into the establishment of the symbiosis during the infection in the larval stage, rather than maintaining the symbiostasis in the trophosome. This genome bridges four decades of physiological research in Riftia, whereas it simultaneously provides new insights into the development, whole organism functions, and evolution in the giant tubeworm.
Keywords: vestimentiferan, host–symbiont interaction, organismal evolution, comparative genomics/transcriptomics
Introduction
The discovery of the giant tubeworm Riftia pachyptila (Jones 1981) at deep-sea hydrothermal vents on the Galapagos Spreading center in 1977 (Corliss et al. 1979) has initiated the onset of a continuous torrent of studies (Childress and Fisher 1992; Nelson and Fisher 1995; Stewart and Cavanaugh 2006; Bright and Lallier 2010; Childress and Girguis 2011; Hilário et al. 2011). With its enormous size (Fisher et al. 1988; Hessler et al. 1988; Shank et al. 1998), rapid cell proliferation (Pflugfelder et al. 2009), seemingly fast growth (Lutz et al. 1994, 2001), but short life (Klose et al. 2015) one of the most puzzling findings was the lack of a digestive system in an animal with a highly unusual body plan (Jones 1981). Descriptions of mouth- and gutless pogonophoran relatives go back a century (Caullery 1914). The first vestimentiferans Lamellibrachia barhami Webb, 1969 and Lamellibrachia luymesi van der Land and Nørrevang, 1975 were described already a few years earlier than Riftia. However, it was the discovery of Riftia, thriving in an apparently poisonous hydrothermal vent environment, which sparked the discovery of the first-described chemosynthetic animal–microbe symbiosis (Cavanaugh et al. 1981); an association in which Riftia, without a mouth or a gut, relies on the sulfide oxidizing chemoautotrophic symbionts for nutrition (Cavanaugh et al. 1981; Felbeck 1981; Arp and Childress 1981, 1983; Rau 1981a, 1981b).
Despite the fact that neither the animal host, nor the symbiont, nor the intact association are amenable to long-term cultivation, Riftia is easily one of the best studied deep-sea animals which have consistently led to major discoveries (reviewed by Bright and Lallier [2010]). Crucial was the development of various devices to measure chemical and physical parameters directly in the deep sea to understand the abiotic conditions under which this tubeworm thrives at vigorous diffuse vent flow (Hessler et al. 1988; Shank et al. 1998; Luther et al. 2001; Le Bris et al. 2003; Mullineaux et al. 2003; Le Bris, Govenar, et al. 2006; Le Bris, Rodier, et al. 2006). Unprecedented and equally important was the development of high-pressure flow-through systems to simulate in situ conditions in the lab (Quetin and Childress 1980; Girguis et al. 2000).There has been probably no deep-sea animal with more resourceful experimental approaches applied in situ and ex situ than Riftia, for example, catheterized tubeworms under flow-through pressure (Felbeck and Turner 1995), artificial insemination and developmental studies under pressure (Marsh et al. 2001), predation experiments with mesh cages in situ (Micheli et al. 2002), hydraulically actuated collection devices of tubeworm aggregations (Hunt et al. 2004; Govenar et al. 2005), artificial plastic tube deployments (Govenar and Fisher 2007), pressurized experiments (Goffredi et al. 1997; Shillito et al. 1999; Girguis et al. 2000, 2002), and finally, various in situ settlement devices for tubeworm larvae (Mullineaux et al. 2000, 2020; Nussbaumer et al. 2006). These innovative experiments associated with four decades of research taught us about many aspects of Riftia’s evolution and biology.
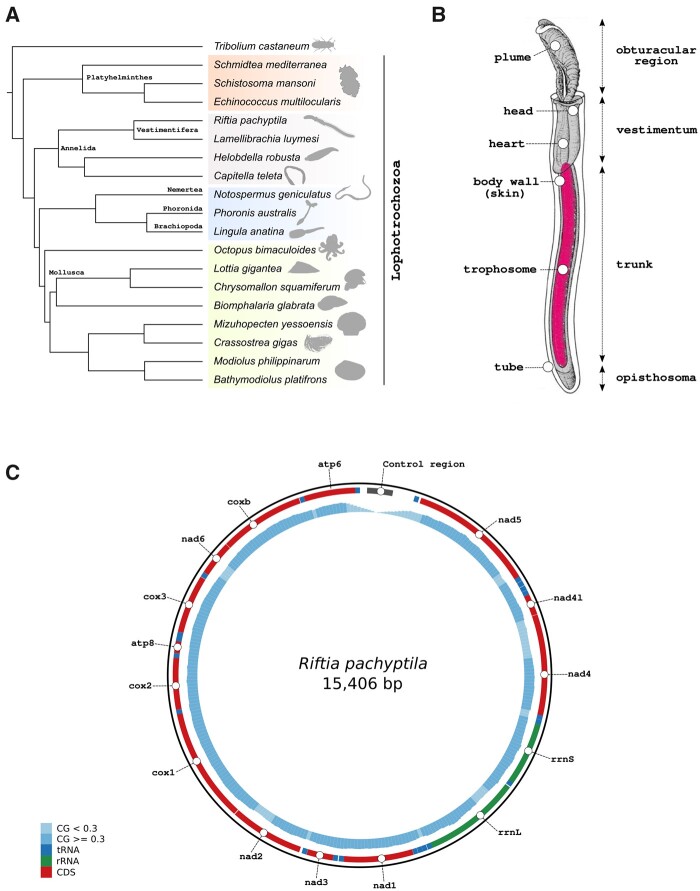
After many microanatomical studies accompanied by heated, highly controversial phylogenetic discussions, the question of who the closest relatives of Riftia are was ultimately solved by traditional cladistic and novel molecular analyses (Fauchald and Rouse 1997; McHugh 1997; Halanych et al. 2001; Rouse 2001; Schulze 2002). They showed that vestimentiferans are lophotrochozoan polychaetae worms within Annelida (fig. 1A) (Polychaeta, Siboglinidae, Vestimentifera) (Pleijel et al. 2009). Similar to many other polychaetes, Riftia is gonochoristic with internal fertilization and undergoes a biphasic life cycle with a pelagic phase including indirect development through spiral cleavage and a trochophore larvae (Marsh et al. 2001). The benthic phase is marked by the uptake of the symbiont into the metatrochophore larvae and growth into an adult, which completely reduces its mouth, gut, and anus. Instead, a unique mesodermal nutritional organ, the trophosome, functionally replaces the digestive system (Nussbaumer et al. 2006; Bright et al. 2013). The adult body is organized into four distinct regions, the obturacular region, the vestimentum, the trunk, and the opisthosoma (fig. 1B). The anterior obturacular region of the animal projects a vascularized branchial plume, which is responsible for the sequestration of nutrients and gas exchange, followed by the vestimentum, a muscular head region enclosing the heart, brain, the excretory organ, and the gonopores. The trunk region, the single elongated first segment, harbors the trophosome and the gonads. The posterior part, the opisthosoma, contains a typical segmented annelid region with serially arranged chaetae (Bright et al. 2013). It is so far unknown how this unusual body plan lacking the entire digestive system is reflected in their developmental genes and signaling pathways. Gutless parasitic tapeworms, for example, have lost many developmental genes including all ParaHox genes (Tsai et al. 2013).
Fig. 1.
Overview of Riftia pachyptila body plan anatomy, phylogenetic placement, and mitochondrial genome. (A) Phylogenetic placement of Riftia pachyptila within Lophotrochozoa. Riftia together with Lamellibrachia form the clade Vestimentifera, a group of marine animals living in chitinous tubes and lacking a digestive tract. Animal silhouettes were downloaded from http://phylopic.org/. Tree topology was obtained through phylogenomic analysis. (B) Schematic drawing of Riftia pachyptila. The first part of the body, the obturacular region, contains the highly vascularized plume, whereas the head, heart, and gonads are located in the second body part, the vestimentum. The trunk region, third body part, harbors the trophosome (organ that houses the symbiotic bacteria), and the body wall (skin). The posterior part, the opisthosoma is the fourth and last body region of the tubeworm. Schematic drawing was modified from Nussbaumer et al. (2006). (C) Schematic representation of Riftia pachyptila mitochondrial genome, including the complete control region. CG-content and tRNA genes are represented by the blue histograms and boxes, respectively.
The trophosome of Riftia, a soft multilobed and highly vascular tissue, houses a polyclonal endosymbiotic population dominated by one genotype of Candidatus Endoriftia persephone, a chemoautotrophic gammaproteobacteria (Robidart et al. 2008; Gardebrecht et al. 2012; Polzin et al. 2019) that oxidizes sulfur compounds via oxygen and nitrate and, in turn, harnesses that energy to fix dissolved inorganic carbon (or DIC, which includes carbon dioxide and bicarbonate) to organic matter. Briefly, the trophosome is far removed, and has no direct contact with external environment, so the host presumably provides all of the inorganic nutrients to the symbionts. This primarily occurs via the highly vascular brachial plume, which takes up oxygen and hydrogen sulfide (H2S) from the external environment and transports these to the trophosome via a complex and unique complement of hemoglobins (Arp and Childress 1981, 1983; Arp et al. 1987; Zal et al. 1996, 1997; Bailly et al. 2002; Flores et al. 2005). DIC is also taken up by Riftia, which is unusual as carbon dioxide is an animal respiratory waste product. However, in this case the worm must provide additional DIC to the symbionts for net carbon fixation, and does so by accumulating DIC in the blood (Goffredi et al. 1997, 1999). Moreover, physiological studies have shown that Riftia also takes up nitrate (also unusual for an animal), and in turn the symbionts reduce it to organic nitrogen (Hentschel and Felbeck 1993; Girguis et al. 2000). In return, the host is nourished through the symbiont releasing organic matter and symbiont digestion, which occurs prior to bacteriocyte death in the periphery of the trophosome lobules (Felbeck 1985; Hand 1987; Felbeck and Jarchow 1998; Bright et al. 2000; Hinzke et al. 2019). Despite four decades of research, key questions about trophosome function remain, including but not limited to: 1) which of the two nutritional modes is more important (organic matter release or symbiont digestion; Bright et al. 2000) and 2) the mechanisms that underlie organic nitrogen synthesis and distribution between the symbionts and the host.
Despite the highly derived annelid body plan, symbiotic lifestyle, and over 40 years of extensive physiological research, whole-genome information of Riftia has been lacking. Here, we generated a high-quality genome draft and distinct tissue-specific transcriptomes of the giant gutless tubeworm Riftia. By analyzing the genome and transcriptomes of Riftia in a comparative framework, we highlight many evolutionary adaptations related to the obligate symbiotic lifestyle and survival in the deep-sea hydrothermal vent environment. The Riftia genome, together with a transcriptome and proteome study (Hinzke et al. 2019), a transcriptome study on the close relative Ridgeia piscesae (Nyholm et al. 2012), the genomic resources available for other close related tubeworms (L. luymesi—short Lamellibrachia; Paraescarpia echinospica—short Paraescarpia) (Li et al. 2019; Sun et al. 2021), and an extensive body of research broadens our understanding of one of the most conspicuous models for host–symbiont interaction and of the biology of Vestimentifera. Most importantly, we show that the developmental gene repertoire is conserved, and that besides the well-known nutritional aspect of the trophosome, its mesodermal origin brought an inherited suite of functions such as, hematopoiesis, endosomal digestion of endosymbionts, and storage of excretory products likely adapted to serve host–symbiont physiological interactions. Although the innate immune system is apparently downregulated in the presence of the symbiont, it is highly active in the remaining body directly exposed, or connected through openings to, to the environment.
Results and Discussion
Riftia Represents the Most Complete Annelid Genome to Date Including a Complete Mitogenome
To assess the whole-genome content of the giant tubeworm (Jones 1981), we sequenced a single individual from the hydrothermal vent site Tica, East Pacific Rise 9°50′N region, with approximately 87-fold coverage using Pacific Biosciences Sequel system (supplementary figs. 1–3 and supplementary table 1, Supplementary Material online). We found the haploid genome size (560.7 Mb with a N50 length of ∼2.8 Mb) to be smaller than previous genome-size estimates (Bonnivard et al. 2009) (table 1 and supplementary fig. 4, Supplementary Material online). The Riftia GC value is 40.49%, and the repeat content accounts for 29.99% of the total length of the genome with most of the repetitive landscape dominated by interspersed and unclassified lineage-specific elements (35.2%) (supplementary fig. 5, Supplementary Material online). After genome postprocessing, we identified a total of 25,984 protein coding genes with homologue, transcriptome, ab initio, and gene expression evidence. The BUSCO4 (Simão et al. 2015) genome completeness score is 99.37%. These numbers render Riftia the most complete annelid genome to date (Simakov et al. 2013; Li et al. 2019; Martín-Durán et al. 2020; Sun et al. 2021) (supplementary fig. 6, Supplementary Material online).
The complete reconstruction of siboglinid mitochondrial genomes including the AT-rich control region has been notoriously difficult (Li et al. 2015). In this case, we were able to obtain it due to deep long read sequencing. The 15,406 bp circular mitochondrial genome contains all expected 13 coding sequence genes, two ribosomal RNA genes, and the 22 tRNAs, typical of bilaterian mitogenomes (fig. 1C and supplementary fig. 7, Supplementary Material online) (Boore 1999). In contrast to two other Riftia reference mitogenomes (Jennings and Halanych 2005; Li et al. 2015), we recovered the full control region (D-loop), yielding a mitochondrial genome longer than those previously reported. The gene order and the number of genes are conserved among all three Riftia and other siboglinids reference mitogenomes, though there are size differences that are most likely due to the incomplete nature of previously published genomes.
The Developmental Gene Repertoire in Gutless Riftia Is Conserved
Because of the lack of molecular information on the development of cell types and the evolution of the vestimentiferan body plan, we identified and annotated a suite of key developmental genes and signaling pathway-related genes in the giant tubeworm genome. We found that key genes involved in the development of the digestive tract in metazoans (Hejnol and Martín-Durán 2015; Nielsen et al. 2018), such as goosecoid, brachyury, foxA, and all three ParaHox genes, xlox, cdx, and gsx, present in the Riftia and Lamellibrachia genomes (supplementary figs. 8–10, Supplementary Material online). The conservation of these genes in vestimentiferans is apparently not only crucial for developmental processes but also serves the microphagous nutrition in settled larvae until nourishment by the symbionts takes over in juveniles (Nussbaumer et al. 2006).
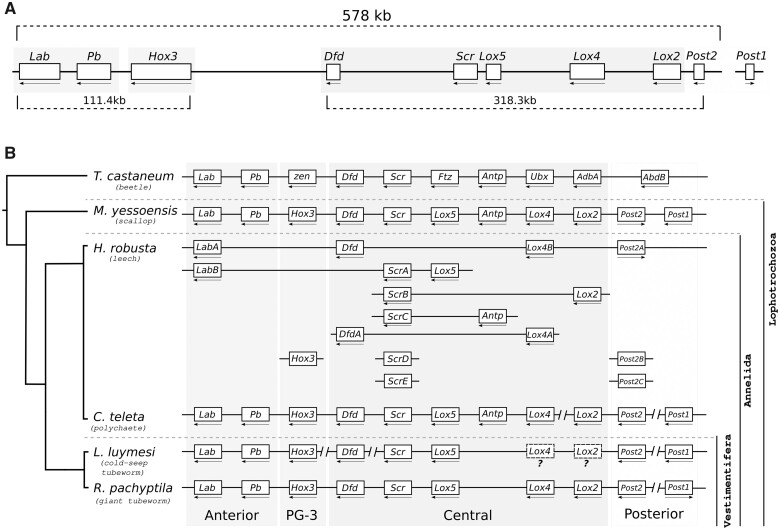
The Hox cluster (∼578 kb in size—fig. 2A), homeodomain-containing transcription factors (TFs) with roles in anterior–posterior axial identity in metazoans (Pearson et al. 2005; Duboule 2007), is nearly intact and complete in the giant tubeworm genome (supplementary figs. 8 and 9 and supplementary note 2, Supplementary Material online). The same complement and synteny were identified in the chromosomal-level genome of Paraescarpia (Sun et al. 2021), attesting the good completeness and contiguity of the Riftia genome. We did not identify hox7 in Riftia, indicating a secondary loss of this gene in the giant tubeworm, a pattern also observed in other lophotrochozoan representatives such as phoronids (Luo et al. 2018) and bivalves (Gerdol et al. 2015; Calcino et al. 2019). Hox7, lox2, and lox5 are missing from Lamellibrachia genome suggesting a possible loss of the central Hox cluster elements (fig. 2B), contradicting recent results (Sun et al. 2021) (but see supplementary note 2, Supplementary Material online). The Hox-like elements, homeotic genes equally important for body plan specification and developmental processes, gbx, evx, mox, mnx, en, and dlx were also found in the giant tubeworm genome. Engrailed (En) and even-skipped (Evx) have two and four copies, respectively (supplementary fig. 8, Supplementary Material online).
Fig. 2.
The Hox gene complement of Riftia pachyptila and selected metazoans. (A) Hox cluster organization in the genome of Riftia pachyptila. Nine out of the ten Hox genes are located in one single genomic scaffold. Hox7 is missing from the giant tubeworm genome. Arrows indicate direction of transcription. Only the longest gene model is shown. (B) Hox cluster present in selected metazoans. Part of the central Hox class is missing from the cold-seep tubeworm Lamellibrachia (but see supplementary note 2, Supplementary Material online). Helobdella and Capitella clusters are based on Simakov et al. (2013), whereas Mizuhopecten cluster is from Wang et al. (2017).
Few signaling pathways are required to control cell-to-cell interactions and produce the plethora of cell types and tissues in Metazoa (Pires-daSilva and Sommer 2003) among them TGFβ, Wnt, Notch, and Hedgehog (Moustakas and Heldin 2009; Ingham et al. 2011; Holstein 2012; Massagué 2012; Niehrs 2012; Gazave et al. 2017) (supplementary figs. 11–15, Supplementary Material online), The Riftia genome contains 14 TGFβ genes, including nodal and its antagonist lefty, the latter previously assumed to be a deuterostome innovation (Simakov et al. 2015). Notch and hedgehog are present as single copy genes in the Riftia genome as well as in Lamellibrachia, however, the notch receptor jagged is missing from both tubeworms. Jagged is present in the annelids Capitella teleta, Helobdella robusta, and Platynereis dumerilii (Gazave et al. 2017), suggesting a secondary loss in Vestimentifera. Patched and dispatched genes, membrane receptors for the hedgehog ligand (Ingham et al. 2011) are present in Riftia with the dispatched genes expanded in vestimentiferans. In Riftia, we identified the 12 expected Wnt ligands (Wnt3 has been shown to be lost in the Protostomia lineage) and their receptors frizzled, smoothened and sFRP (Holstein 2012). There is a genetic linkage of Wnt1, 6, 9, and 10 in Riftia akin to the gastropod Lottia gigantea and the fruit fly Drosophila melanogaster (Cho et al. 2010), reaffirming the ancient protostomian ancestral conserved linkage. The remaining eight Wnt genes in Riftia are disorganized on eight different scaffolds. Overall, despite the highly derived body plan, Riftia presents a deep conservation of the developmental gene toolkit akin to many distinct bilaterian animals.
The Riftia Genome Is Characterized by Reductive Evolution
Multiple lines of evidence point to a relatively small genome, with gene family contractions exceeding expansions in Riftia, indicative of reductive evolution. The giant tubeworm genome is approximately 168 Mb smaller than Lamellibrachia, its relative from cold hydrocarbon seeps whose genome is approximately 688 Mb with a N50 of 373 kb (Li et al. 2019). The difference can be attributed to the increased number of repeat elements and protein coding genes in the cold seep tubeworm (38,998 gene models and repetitive content of 36.92%; supplementary note 1 and supplementary fig. 16, Supplementary Material online).
To identify clusters of orthologous genes shared among the two vestimentiferans Riftia and Lamellibrachia, the polychaete C. teleta (herein called Capitella), and the clitellid Helobdella robusta (herein called Helobdella) (Simakov et al. 2013), we employed tree-based orthology inferences (Emms and Kelly 2019). The annelid core genome, the collection of orthogroups shared among the four annelids, contains 6,349 cluster of orthologous genes. Less than half of them represent the vestimentiferan core genome (2,883 orthogroups) shared between Riftia and Lamellibrachia. Interestingly, the number of shared orthogroups between the Riftia-Helobdella (17) and -Capitella (116) pairs are smaller than those between Lamellibrachia-Helobdella (89) and -Capitella (349) pairs, indicating that Riftia contains a more derived gene repertoire than its close relative Lamellibrachia.
To further investigate the important processes of gene losses and gains, known to shape animal evolution (Fernández and Gabaldón 2020; Guijarro-Clarke et al. 2020), identify expanded protein domains, taxonomically restricted genes, and positively selected genes in Riftia, we employed multilevel comparative approaches involving statistical analysis, taxon rich orthology inferences (N = 36), and sensitive similarity searches (supplementary table 2 and supplementary note 3, Supplementary Material online). The Riftia genome shows a net reduction of gene numbers with only 734 expanded but 1,897 contracted gene families, whereas the evolutionary history of Lamellibrachia is characterized by gene gains. Notably, the average expansion value of gene families in Riftia is the lowest among the four selected annelids herein analyzed (supplementary fig. 17, Supplementary Material online). A total of 8,629 lineage-specific genes (∼33.21% of the total) were identified in Riftia. Compared with the giant tubeworm, Lamellibrachia contains more lineage-specific genes (10,262–26.31% of the total).
The contracted gene families are not restricted to any specific biological process, as revealed by our gene ontology (GO) term enrichment analysis (N = 18) (supplementary fig. 18, Supplementary Material online). Rather it appears that the giant tubeworm genome is undergoing a broad reduction in gene content. Among the contracted gene families are genes controlling the transcriptional machineries (supplementary fig. 19 and supplementary table 3, Supplementary Material online). TFs are proteins with sequence-specific DNA-binding domains that control gene transcription and tissue identity (Schmitz et al. 2016). To gain understanding into the repertoire of TFs in Riftia, we annotated and classified genes in the tubeworm genome present in five major groups of TFs (bzip, homeobox, nuclear factor, bHLH, and zinc-finger) with sensitive similarity searches. The giant tubeworm presents the lowest number of TFs within the analyzed annelids (414), supporting our gene family analysis (discussed below). The cold-seep tubeworm genome contains a similar complement size as Riftia (423), with Capitella (551) and Helobdella (568) presenting a higher number of TF genes, comparatively. These results point to pervasive TF losses in the Vestimentifera lineage (supplementary note 3, Supplementary Material online).
Expanded and Lineage-Specific Gene Families in Riftia
Despite overall genome reduction, the Riftia genome exhibits, there is also a variety of expanded gene families (supplementary note 3, supplementary fig. 20, and supplementary table 4, Supplementary Material online). These expanded families are enriched with GO terms associated with sulfur metabolism, membrane transport, and detoxification of xenobiotic, for example, foreign substances (xenobiotic transmembrane transporter activity, galactosylceramide sulfotransferase activity, CoA-transferase activity) (Gamage et al. 2006), detoxification of hydrogen peroxide as antioxidative stress response (glutathione catabolic and biosynthetic processes) (Espinosa-Diez et al. 2015), neurotransmitter- and ion channel-related functions (sodium symporter activity), oxygen transport (oxygen binding, hemoglobin complex), endosomal degradation (lysozyme activity), and secretion of chitin (chitin binding, protein glycosylation) (discussed with more details later).
Genes involved in the production of extracellular components of vestimentiferans such as the cuticle and the basal matrixes as well as the tube and chaetae (Gardiner and Jones 1994) were found in expanded families of Riftia (as well as Lamellibrachia), some of which are specific to either Riftia or Lamellibrachia (supplementary note 3, supplementary figs. 21–24, and supplementary tables 5 and 6, Supplementary Material online). Additionally, integrated genomic, transcriptomic, and proteomic analyses in Paraescarpia revealed a fairly similar scenario (Sun et al. 2021), pointing to common and shared molecular mechanisms involved with tube formation in cold-seep and vent vestimentiferans.
The Riftia genome contains expanded protein domains related to several high-molecular mass proteins such as laminin, nidogen, and collagen. These proteins are part of extracellular matrix secreted basally from epithelia, also known to regulate cellular activity and growth in other animals (Timpl and Brown 1996). In Riftia, extensive short collagen fibers are found below the epidermis, extending between muscles cells, and building the matrix of the obturaculum. In addition, long helically arranged collagen fibers are the main component of the cuticle apically secreted from the epidermis (Gardiner and Jones 1994). Importantly, many genes involved in chitin production, a biopolymer part of the hard protective tube secreted from pyriform glands of the vestimentum, trunk, body wall, and opisthosoma (Gardiner and Jones 1993), are taxonomically restricted to the Riftia lineage. Expectedly, we identified in the vestimentum and body wall tissues of Riftia several tissue-specific genes (TSGs) involved in the chitin metabolism responsible for the tube production as well as dissolution (supplementary figs. 25 and 26, Supplementary Material online). Although the specific gland type responsible for dissolution of tube material has yet to be identified, we suggest that in Riftia the straight tube, that can reach up to 3 m in length and 5 cm in diameter (Gaill and Hunt 1986; Grassle 1987; Fisher et al. 1988), can only widen in diameter to accommodate growth of the worm when tube material is dissolved and newly secreted, which agrees with the distribution of many TSG involved with tube biosynthesis. Overall, our findings of these expanding gene families as well as gene expression patterns underline the importance of chitin in Riftia, which is considered one of the fastest growing invertebrates (Lutz et al. 1994, 2001). In order to achieve these high growth rates, Riftia needs to both digest and remodulate its own tube with astonishing speed.
Furthermore, the multilevel comparative analyses revealed an enrichment of GO terms in the lineage-specific Riftia genes involved with the control of the chromosome condensation and nucleosome assembly, and positively selected genes related to tumor suppression (PIN2/TERF1-interacting telomerase inhibitor) and transcription initiation (TFIIB- and -D) (Roeder 1996; Zhou and Lu 2001). Interestingly, in Lamellibrachia smad4 (Li et al. 2019), which is a tumor suppressor and TF, is under positive selection, suggesting a common vestimentiferan evolutionary adaption responsible for controlling the chromatin-remodeling events and the extraordinarily cell proliferation rates in these two tubeworms (supplementary table 7 and supplementary note 3, Supplementary Material online) (Pflugfelder et al. 2009).
The protein annotation of the rapidly evolving expanded gene families in Riftia identified members of the complement system involved in innate immunity and self-, nonself-recognition (sushi repeat domain-containing protein) (Kirkitadze and Barlow 2001). Riftia contains the greatest number of sushi-domain-containing proteins among lophotrochozoans, presenting a total of 42 copies which are organized either in genomic clusters or dispersed as single elements throughout the genome (supplementary figs. 4 and 27, Supplementary Material online). Of these, only 40 are shared with the cold seep tubeworm Lamellibrachia, pointing to a lineage-specific expansion at the base of Vestimentifera. Sushi genes, a common component of hemocytes (i.e., immune cells with phagocytic function; Pila et al. 2016), have been implicated in the mediation of the host–symbiont tolerance in the bobtail squid Euprymna scolopes—bioluminescent Aliivibrio fischeri association (McAnulty and Nyholm 2017). Although the rapid evolution of these proteins in Riftia and Lamellibrachia suggests similar evolutionary adaptations to the tubeworm/endosymbiont mutualism, the absence of any significant expressions in adult tissues rather point to their involvement in recognition of the symbiont during transmission in the larval stage or to potential pathogen recognition upregulated upon exposure.
Substrate Transport for Energy Conservation and Biosynthesis Is Supported by Lineage-Specific Adaptations and Parallel Evolutionary Events in Riftia
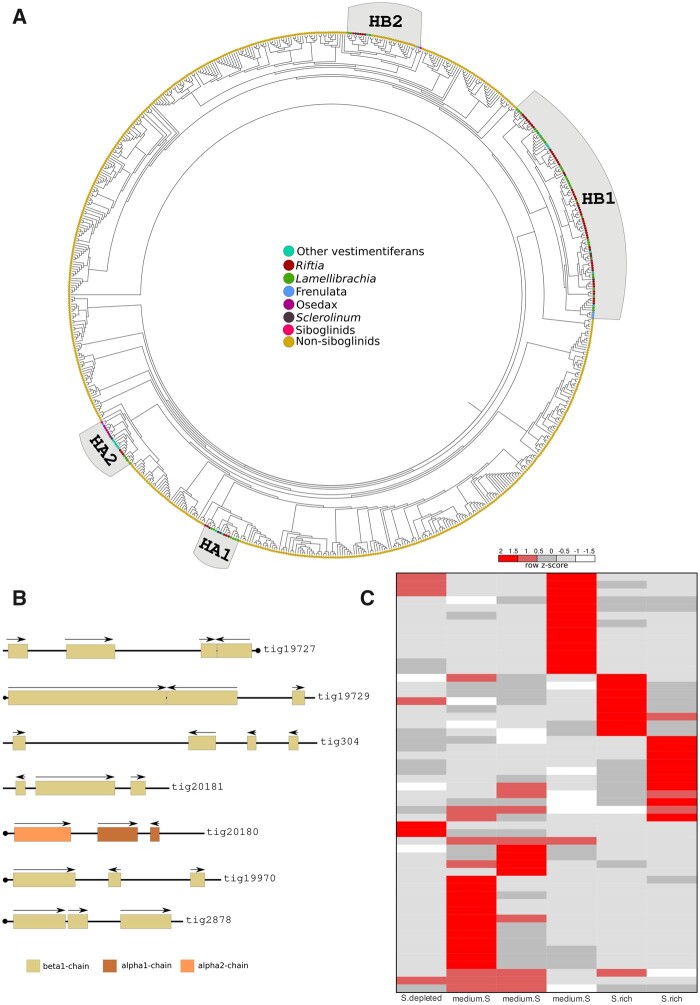
As an adaptation to the sulfidic vent environment, and in support of a symbiotic lifestyle, the respiratory pigments in Riftia, and other vestimentiferans such as Lamellibrachia, bind noncompetitively and reversibly to oxygen and sulfide, simultaneously providing a key substrate for chemosynthesis by the symbionts while also averting the sulfidic inhibition of the hosts’ mitochondrial oxidative chain reactions (Arp and Childress 1983; Terwilliger et al. 1985). Our previous gene family evolution analysis identified an expansion of hemoglobins in the giant tubeworm genome compared with other nonvestimentiferan lophotrochozoans (supplementary fig. 21 and supplementary table 6, Supplementary Material online and fig. 3A). Additionally, a recent genome study found a massive expansion of β1-hemoglobin in the cold seep tubeworm Lamellibrachia (Li et al. 2019). To gain better insights in the evolution of Hb and linker genes in the Vestimentifera lineage, we employed thorough comparative genomics, phylogenetics, domain composition, and gene quantification analyses. The genomic arrangement of the giant tubeworm Hb genes indicates that they were originated through a series of tandem duplications, totaling seven distinct genomic clusters (fig. 3B). We annotated 26 extracellular Hbs and six linker genes in the Riftia genome (supplementary figs. 28 and 29 and supplementary note 4, Supplementary Material online). Twenty-two Hb genes were phylogenetically placed in the β1-Hb group, surpassing previous estimates of the β1-Hb complement in the giant tubeworm (Bailly et al. 2002; Sanchez et al. 2007; Hinzke et al. 2019). α2- and β2-Hbs are found as single copy genes, whereas α1-Hb group contains two paralogous genes. The sulfide-binding ability of the Riftia Hbs is associated with the occurrence of free cysteine residues in one α2 and one β2 Hb genes (Bailly et al. 2002), as well as the formation of persulfide groups on linker chains (Zal et al. 1998; Bailly et al. 2002). Our results show that seven additional paralogous genes belonging to the β1-Hb group contain the putative free-cysteine residues, which were confirmed through multiple sequence alignments and homology model generation (supplementary figs. 30 and 31 and supplementary note 4, Supplementary Material online). Additionally, it has been hypothesized that zinc ions, rather than free-cysteine residues, are responsible for the H2S binding and transport on vestimentiferan α2 chains (Flores et al. 2005). We identified the three conserved histidine residues (B12, B16, and G9), predicted to bind zinc moieties, in Riftia Hb genes. However, we observed variations within the Lamellibrachia α2 genes. A broader comparison of α2-Hb genes belonging to different annelid taxa challenged the hypothesis of zinc sulfide-binding mechanisms for H2S in siboglinids and vestimentiferans (Li et al. 2019). Our results, solely based on the conservation of histidine residues, corroborate Flores et al. (2005) hypothesis that zinc residues may be involved in the sequestration and transport of hydrogen sulfide at least on the giant tubeworm.
Fig. 3.
Expanded hemoglobin complement in Riftia pachyptila. (A) Midpoint rooted phylogeny of 693 Riftia, annelid and metazoan hemoglobin genes, using Belato et al. (2019) as backbone. Colored circles correspond to different annelid taxa and metazoans. (B) Seven genomic hemoglobin clusters are present in the Riftia genome. Arrows indicate the direction of transcription. Scaffolds with a circle on the end indicate the presence of Hb genes in the terminal end of the genomic segment. Only the longest gene models are shown. Colors represent the different hemoglobin chains. (C) Heat map expression of hemoglobins in the trophosome under three experimental conditions (obtained from Hinzke et al. [2019]): medium sulfide (medium.S), sulfide rich (S.rich), and sulfide depleted (S.depleted).
To investigate the gene expression dynamics of the newly and previously identified Hb paralogs in Riftia, we analyzed published transcriptomes sampled from Riftia’s trophosomes containing sulfur-rich to sulfur-depleted symbionts (Hinzke et al. 2019). Hb gene expression showed great variation, indicating a more specialized role of the Hbs according to the environmental chemical fluctuations in the unstable deep-vent ecosystem (fig. 3C and supplementary note 4 and supplementary table 8, Supplementary Material online). Taken together, these results suggest a more complex system coordinating oxygen-sulfide sequestration and distribution in the giant tubeworm tissues. The Hb complement of Riftia and Lamellibrachia is similar and unique among annelids and lophotrochozoans, in respect to gene numbers and distribution, indicating a Vestimentifera synapomorphy.
As the endosymbionts require carbon dioxide (CO2) for fixing inorganic carbon, the transport of CO2 and the conversion of its alternative forms (e.g., bicarbonate; HCO3−) is mediated by another class of enzymes, the carbonic anhydrases (CAs) (Shively et al. 1998; Cian et al. 2003). We found ten CA genes in the Riftia genome, from which seven are tandemly arrayed in two genomic clusters (supplementary fig. 32, Supplementary Material online). A similar CA complement in Riftia (nine genes) was found in a recent study (Hinzke et al. 2019). To better understand the diversity of CA genes, we analyzed tissue-specific transcriptomes and found at least five CA genes are membrane bound with three of them moderately/highly expressed in the trophosome, indicating that HCO3− conversion to CO2 and diffusion across the bacteriocyte membrane might be a common process in the trophosome, as suggested previously (Sanchez et al. 2007; Bright and Lallier 2010; Hinzke et al. 2019). Tandem duplications and tissue-specific CA expression linked to the intracellular supply of CO2 to endosymbionts have been recently reported in deep-sea bivalves (Ip et al. 2021), showing remarkable resemblance to our findings. Taken together, our results show that the transport of essential compounds to the chemoautotrophic endosymbionts and the maintenance of the mutualistic relationship are driven by lineage-specific and parallel evolutionary events.
Trait and Gene Losses Are Compensated by the Endosymbionts
The loss of the digestive system requires nourishment through the symbiont. The mechanisms of carbon transfer between the endosymbiont and Riftia were shown to be through the fast release of fixed carbon from the symbiont and uptake into host tissue, as well as, through symbiont digestion prior death of the bacteriocytes (Felbeck 1985; Hand 1987; Felbeck and Jarchow 1998; Bright and Lallier 2010; Hinzke et al. 2019). We found corroborating evidence for the uptake of released organic carbon from the symbiont based on the enrichment of GO terms and tissue specificity of succinate-semialdehyde complex genes and nuclear-encoded proteins of the inner mitochondrial membranes (including the tricarboxylate mitochondrial carrier responsible for the transport of succinate) (Majd et al. 2018) in the trophosome (supplementary fig. 33 and supplementary table 9, Supplementary Material online). These results suggest an increased movement of cytosolic succinate through the mitochondrial membrane, possibly increasing the ATP production via the oxidative metabolism. These findings corroborate previous findings and support the involvement of this molecule for nourishment in Riftia from its endosymbiont (Felbeck and Jarchow 1998).
Evidence of digestion was revealed with tissue-specific transcriptome analyses which allowed us to identify the genes involved in the successive stages of lysosomal-associated degradation of symbionts (supplementary fig. 33 and supplementary table 8, Supplementary Material online). The expression of genes associated with endosomal activity, the expression of several lysosomal-associated hydrolases (supplementary figs. 34 and 35, Supplementary Material online), vacuolar ATPases, and small Ras-related GTPases (rab genes) (supplementary fig. 36 and supplementary note 5, Supplementary Material online) in the trophosome, is indicative of lysosomal-associated degradation of symbionts, as suggested earlier in ultrastructural studies, which describe the presence of primary lysosomes and symbionts in different lytic stages in Riftia (Bright et al. 2000; Bright and Sorgo 2005; Hinzke et al. 2019). The high expression of cathepsins and/or legumains in the trophosomes of the cold-seep tubeworms Lamellibrachia and Paraescarpia (Li et al. 2019; Sun et al. 2021) strongly indicates that endosymbiont lysosomal digestion route is widespread in Vestimentifera. In addition, we detected specific genes in the trophosome which are associated with actin cytoskeleton dynamics (ARP2/3) known to be essential for endosomal dynamics (Kast and Dominguez 2017) (supplementary fig. 33 and supplementary table 9, Supplementary Material online). Recent de novo tissue-specific transcriptomes and gene expression quantification of the host and symbiont support the digestive route of nutrition in Riftia/Endoriftia symbiosis (Hinzke et al. 2019).
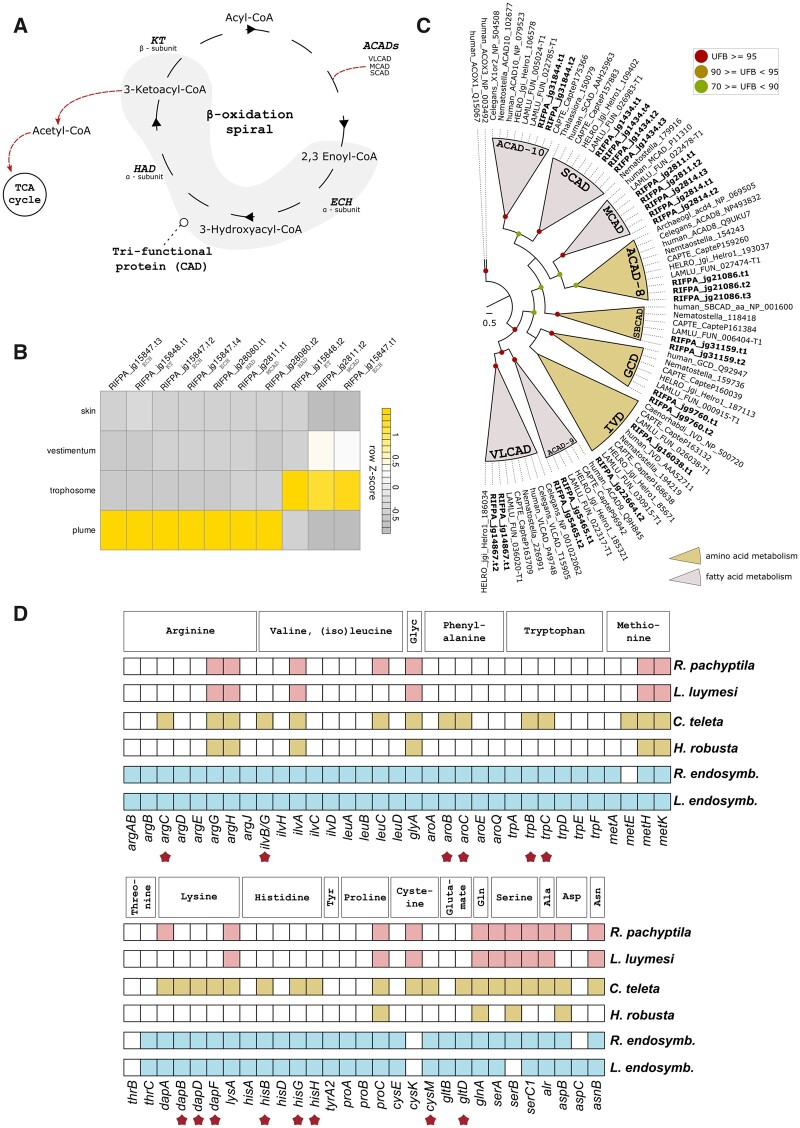
Furthermore, genes involved in the transport of fatty acyl units into the mitochondrial matrix (e.g., carnitine/acylcarnitine carrier) (Indiveri et al. 2011), tricarboxylic acid (TCA) cycle, oxidative phosphorylation, antioxidant systems (i.e., superoxide dismutase II genes and methionine sulfoxide reductases) (supplementary fig. 38, Supplementary Material online), and key players of the fatty acid β-oxidation (FAO) (fig. 4A–C), showed tissue specificity in the trophosomal tissue (supplementary table 9 and supplementary note 5, Supplementary Material online). FAO is a central and deeply conserved energy-yielding process that fuels the TCA cycle and oxidative phosphorylation (Houten et al. 2016). As Riftia relies solely on its endosymbionts for sustenance, the metabolism of fatty acids in the trophosome is certainly linked to the bacterial digestion in this tissue, which is corroborated by a previous proteomic study (Hinzke et al. 2019). Altogether, the results point to different modes of nutrient transfer in the trophosome involving the translocation of released nutrients from symbiont to host through succinate, and the digestion of the symbionts by lysosomal enzymes followed by the degradation of fatty acids using the mitochondrial β-oxidation pathway.
Fig. 4.
Amino acid and fatty acid biosynthesis in Riftia pachyptila. (A) A schematic representation of mitochondrial FAO. The fatty acid degradation is performed in four enzymatic steps involving the membrane bound mitochondrial trifunctional protein and acyl-CoA dehydrogenases. The resulting acetyl-CoA is further oxidized in the TCA cycle. (B) Expression profile of FAO genes. Color coding reflects the expression patterns based on row Z-score calculations. (C) Maximum-likelihood phylogenetic tree inference of the ACAD genes using 1,000 rapid bootstrap replicates. The branch support values are represented by the colored circles in the tree nodes. Accession numbers for NCBI database are displayed after the species names and homologs were retrieved from a previous study (Swigoňová et al. 2009). Capitella, Helobdella, and Lamellibrachia gene identification are derived from the publicly available annotated genomes. (D) Key enzymes related to amino acid biosynthesis identified in Riftia, selected annelids and two tubeworm endosymbiont genomes. The identification of the genes was performed with KEGG and orthology reconfirmed with similarity searches against the ncbi-nr protein database. Riftia and Lamellibrachia lack many amino acid biosynthesis genes indicating nutritional dependence on their endosymbionts. Stars represent genes present in the free-living polychaete Capitella and Endoriftia but absent in Riftia (based on Li et al. [2019] and Tokuda et al. [2013] schemes).
It has been previously described that deep-sea tubeworms are reliant on their endosymbionts for nutrition (Li et al. 2019; Yang et al. 2020). To further explore the nutrient interdependence of Riftia and its endosymbiont, we screened the genome of giant tubeworm and selected annelids for key enzymes related to amino acid biosynthesis (supplementary table 10, Supplementary Material online). We found that Riftia, together with cold-seep tubeworm Lamellibrachia and the parasitic leech Helobdella, lacks many key enzymes related to amino acid biosynthesis when compared with close free-living polychaete relative Capitella (fig. 4D). The identified key enzymes in the giant tubeworm are involved in the biosynthesis of at least 14 amino acids (fig. 4D) similar to Paraescarpia and Lamellibrachia hosts (Li et al. 2019; Yang et al. 2020), whereas Endoriftia possesses the metabolic capability of synthesize all essential and nonessential amino acids. The complete gene toolkit for amino acid biosynthesis is also found in the Lamellibrachia endosymbiont (Li et al. 2019; fig. 4D) but it is more reduced in the endosymbiont of Paraescarpia (which lacks the ability to synthesize threonine and tyrosine) (Yang et al. 2020). Genes involved with amino acid biosynthesis are constitutively expressed across the giant tubeworm tissues, with enzymes related to arginine and glycine metabolism highly expressed in the trophosome (supplementary fig. 39, Supplementary Material online). These findings suggest that loss of key enzymes in mutualistic vestimentiferans as well as a parasitic leech may be due to the beneficial and parasitic relationships, respectively allowing for compensated gene loss, compared with free-living polychaetes.
Overall, endosomal-associated digestion of endosymbionts seems to be a hallmark of intracellular digestion accomplished in the mesodermal trophosome of vestimentiferans, such as Riftia, Lamellibrachia, and Paraescarpia (Nussbaumer et al. 2006; Hinzke et al. 2019; Li et al. 2019; Sun et al. 2021) (see also below). This process serves the host nutrition as well as the control of the symbiont population density during host growth, known from many other symbioses (Angela E. Douglas 2010). In addition, the symbiont provides the host with released organic carbon. Although we do not know yet which partner controls this mode of nutritional translocation, both the evolutionary adaptation of endosymbiont digestion in a mesodermal tissue as well as carbon release contributed to trait loss in one partner compensated by the other (Ellers et al. 2012), and consequently has made Riftia obligatorily associated with its symbiont.
Hematopoiesis Operates in the Trophosome of Riftia
Hematopoiesis, the production of blood cells and pigments, is still a poorly understood process in vestimentiferans. The heart body, a mesodermal tissue in the dorsal blood vessel of vestimentiferans has been hypothesized to be the site of hemoglobin biosynthesis (Schulze 2002). The presence of many TSGs in the trophosome related to 5-aminolevulinate synthase, porphyrin metabolism, and metal ion binding indicate that this tissue harbors the enzymatic machinery necessary for heme biosynthesis. Heme is an integral part of hemoglobin molecules, which is synthesized in a seven multistep pathway that begins and ends in the mitochondrion. To fully characterize the heme biosynthesis pathway in the giant tubeworm, we screened the Riftia genome for the presence of the seven universal enzymes required to synthesize the heme (supplementary fig. 40 and supplementary note 5, Supplementary Material online). The giant tubeworm contains all the seven enzymes present as single copy in its genome with recognizable orthologs in the annelids Lamellibrachia, Capitella, and Helobdella. Gene expression analysis showed that the key enzymes present in the heme biosynthetic pathway are moderately/highly expressed in the trophosome, supporting the GO enrichment analysis (supplementary figs. 40 and 33, Supplementary Material online). The heme biosynthesis in the trophosome is further corroborated by the presence of TSGs in this tissue related to phosphoserine aminotransferase and the mitochondrial coenzyme A transporter, which act as an important cofactor in the final step of heme synthesis and in the transport of coenzyme A into the mitochondria, respectively (Schneider et al. 2000; Fiermonte et al. 2009). These findings confirm the involvement of the mesodermal trophosome in hemoglobin metabolism and suggest that this organ is the site hematopoiesis. In the frenulate Oligobrachia mashikoi, the visceral mesoderm also strongly expresses globin subunits based on in situ hybridization and semiquantitative RT–PCR (Nakahama et al. 2008), but in this siboglinid, the visceral mesoderm is organized as simple peritoneum surrounding the endodermal trophosome (Southward 1993).
Excretory Products Are Stored in the Trophosome of Riftia
The finding of TSGs in the trophosome related to the biosynthesis of nitrogen-containing compounds and in the transport of ornithine (supplementary tables 9 and 11, Supplementary Material online) agrees with the high levels of uric acid and urease activity in this host tissue, as previously reported (Cian et al. 2000; Minic and Hervé 2003).To explore the nitrogen metabolism pathways in Riftia, we identified and quantified the gene expression of several enzymes related to the purineolytic/uricolytic, purine/pyrimidine, taurine, and the polyamine pathways, as well as the urea and ammonia cycles (supplementary figs. 41–45 and supplementary note 6, Supplementary Material online).
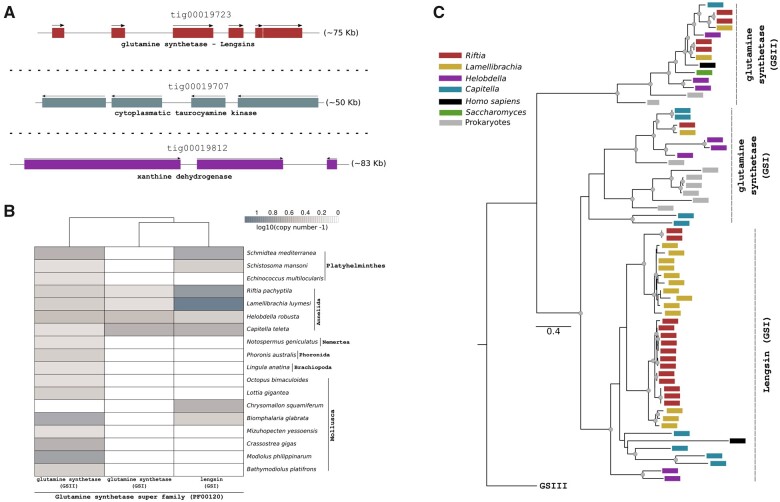
Most of the identified genes are found as single copy in the giant tubeworm genome (supplementary note 6 and supplementary figs. 41–45, Supplementary Material online); however, we identified the presence of three chromosomal clusters harboring glutamine synthetase, cytoplasmatic taurocyamine kinase, and xanthine dehydrogenase/oxidase genes (fig. 5A). These enzymes are involved in the ammonia, urea, and uricolytic pathways, respectively. Riftia and Lamellibrachia contain the highest number of glutamine synthetase genes in the herein investigated lophotrochozoan genomes (fig. 5B). Seven out of the nine glutamine synthetase genes present in Riftia belong to the group I and the remaining to the group II (fig. 5C). Interestingly, only annelid orthologs are found to be phylogenetically close to the prokaryotic group I, indicating a secondary loss of this genes in the remaining lophotrochozoan lineages (see supplementary fig. 43, Supplementary Material online, for the expanded version of the phylogenetic tree). An expanded set of lengsin genes, an ancient class I glutamine synthetase family (Wyatt et al. 2006), is present in Vestimentifera (seven copies in Riftia and 13 in Lamellibrachia). Some members of the newly identified lengsins are also highly expressed in the trophosome, suggesting that these enzymes might play a role in mitigating toxicity of urea, ammonia, and other nitrogenous compounds (Wyatt et al. 2006).
Fig. 5.
Genomic clusters of important genes related to the nitrogen metabolism in Riftia pachyptila, and phyletic distribution and phylogeny of glutamine synthetase genes among selected metazoans. (A) Genomic organization of important genes related to nitrogen metabolism in Riftia. Arrows indicate the direction of transcription. Only the longest gene models are shown. (B) Distribution of glutamine synthase-related genes in the giant tubeworm, closely related annelids, and selected lophotrochozoans. (C) Maximum-likelihood phylogenetic tree inference of members of the glutamine synthetase superfamily using 1,000 ultrafast bootstrap replicates. Colored boxes correspond to different annelids, vertebrates, yeast, and prokaryotes.
The identification of five cytoplasmatic taurocyamine kinase genes, four organized in a genomic cluster (fig. 5A), surpasses previous reports (in which only one cytoplasmatic gene was identified; supplementary fig. 44, Supplementary Material online) (Uda et al. 2005). In accordance with a recent study (Hinzke et al. 2019) and contrasting previous biochemicals investigations on the de novo pyrimidine and polyamine biosynthesis in Riftia (Minic et al. 2001; Minic and Hervé 2003), we identified the trifunctional CAD protein in the genome of the tubeworm reinforcing the notion that Riftia can catalyze the first steps of the pyrimidine synthesis independently of its endosymbionts (supplementary note 6, Supplementary Material online). These results are not unforeseen, since during the aposymbiotic phase (i.e., Riftia’s fertilized egg until the settled larva) pyrimidine metabolism plays a fundamental role in the development and growth of the animal.
We also found that the key enzymes of the uricolytic pathway and urea cycle are highly active in the trophosome (supplementary fig. 41 and supplementary table 8, Supplementary Material online). These results are consistent with enzymatic/light micrograph studies, which show that the trophosome contains high concentration of ammonia, urea, creatinine, and uric acid crystals in the periphery of the lobules (Cian et al. 2000), and with a more recent transcriptomic and metaproteomic study (Hinzke et al. 2019). Surprisingly, we only identified in the complete and closed (de Oliveira AL, in review) and previous endosymbiont genome drafts the subunit-A of the urea transporter (urtA), with the four remaining subunits missing (urtBCDE) (Veaudor et al. 2019). Since all five subunits are required for a proper function of the urea transporter, these results challenge the idea of an active shuttle of urea from the host to the endosymbiont (Robidart et al. 2008).
Cell Proliferation and Cell Death Interplay with Innate Immunity in Riftia
To better understand how fast growth (Lutz et al. 1994) fueled by high proliferation rates (Pflugfelder et al. 2009) and innate immunity act in Riftia in tissues exposed to the environment and in the endosymbiont-housing trophosome, we characterized the key molecular components, and their gene expressions, of important pathways related to cell cycle signaling (supplementary fig. 46, Supplementary Material online), Toll-like receptor/MyD88 (supplementary figs. 47–49, Supplementary Material online), as well as the apoptotic (supplementary figs. 50–57, Supplementary Material online) and autophagic (i.e., macroautophagy) cell death events (supplementary fig. 58, Supplementary Material online). The Riftia genome similar to other investigated lophotrochozoans and closely related annelids harbors all the key components of these conserved pathways (supplementary note 7, Supplementary Material online) (Zhang, Fang, et al. 2012; Sun et al. 2017; Luo et al. 2018; Li et al. 2019; Ip et al. 2021). We did not identify any extensive remodeling (i.e., gene family expansions and contractions) of the immune (with the exception of sushi genes) and programed cell death components in the giant tubeworm genome, as shown to be important in the maintenance of host–symbiont interactions in deep-sea mussels and clams (Sun et al. 2017; Ip et al. 2021).
Overall, Riftia’s gonad and plume tissues are highly active in cell proliferation and programed cell death. Subject to potential pathogen infections through the gonopore opening and direct contact to the vent water (Jones 1981), respectively, these tissues show the entire suite of genes involved in the innate immunity recognition with TLRs, downstream cellular immune responses, as well as apoptosis, autophagy, and endosomal-related genes. These results were additionally supported by the GO enrichment analyses in the female gonad and plume tissues (supplementary figs. 59 and 60, Supplementary Material online). The trophosome, in contrast, despite the remarkably high bacterial population density (Powell and Somero 1986; Bright and Sorgo 2005) does not show any striking upregulation of TLR for endosymbiont recognition, nor cell proliferation, nor programed cell death pathways (at least not in the classical sense; see Hinzke et al. 2019). Instead, we found few moderately/highly expressed genes present in the immune system (irak2 and 4, tab1, tak1, mkk3/6), cell cycle (cyclin A, B2, D2, cdk4), apoptotic (cas2, cas8, birc8), and autophagic (becn1, atg2b-7-8-16) pathways in the trophosome of Riftia. Interestingly, a previous study suggested that immune-related genes were significantly more expressed in the trophosome in relation to other symbiont-free tissues in the siboglinid Ridgeia piscesae, positing a more important role of the immune system in the host-endosymbiont homeostasis (Nyholm et al. 2012).
Few other individual components of the innate immune system, that is, bactericidal permeability-increasing proteins and pattern recognition receptors, have been implicated in symbiont population control in tubeworms (Nyholm et al. 2012; Hinzke et al. 2019). However, based on our broad gene expression analyses, we argue that the host immune system does not play a major role in taming the endosymbiont population in the trophosome, as previously suggested (Hinzke et al. 2019). Furthermore, immunohistochemical and ultrastructural cell cycle analyses identified apoptotic and proliferative events in the trophosome (Pflugfelder et al. 2009), indicating that despite the overall low expression of gene markers related to these pathways described herein, these events occur in this tissue. In which extent these different pathways interact to shape the host/symbiont interactions and to maintain tissue homeostasis remains to be shown, however, it is clear that multiple and not mutually exclusive programed cell-death, immune-related, and proliferative events (supplementary note 7, Supplementary Material online) are acting on the trophosome.
From Phenotype to Genotype and Back
After 40 years of intensive research, we are now finally able to integrate the obtained genome and tissue-specific transcriptome information with the current body of knowledge on the phenotype to better understand the genotype–phenotype interplay in the giant tubeworm. The R. pachyptila genome is characterized by reductive evolution with broad gene family contractions exceeding gene family expansions. Compared with the close relative L. luymesi (Lamellibrachia live at longer-lived and less physiologically taxing hydrocarbon seeps), Riftia exhibits a more derived gene repertoire for important traits related to symbiosis and the highly disturbed and stressful hydrothermal vent habitat they inhabit in the deep sea.
The mutualism between Riftia and its symbiont has not transited from individuality of symbiotic partners to a new integrated organism (Szathmáry and Smith 1995) because it lacks mutual dependency (Kiers and West 2015; West et al. 2015). Riftia is, in fact, one of the few examples known in which dependency is asymmetric with a facultative horizontally transmitted symbionts, which have the capacity to live with or without the host. The Riftia host, however, is obliged to partner with the symbiont or else they cannot thrive. Therefore, the host’s fitness is strictly tied to the persistence of this association over ecological and evolutionary time scales. The genome data now clearly show the peculiarities and divergencies in Riftia’s genotype compared with closely related free-living annelids and other lophotrochozoans, as well as which evolutionary adaptations of the host genotype ensure the maintenance of the association.
We found that despite the drastic morphological remodeling during its early development leading to the mouth-, gutless adult animal, Riftia retained the highly conserved developmental gene repertoire present in other lophotrochozoans and distant related animals. These results can be interpreted as counterintuitive considering that the adult body plan alone provides little unambiguous evidence of the vestimentiferan phylogenetic relationship. These animals were initially compared with deuterostomes (Caullery 1914) and considered related to hemichordates (Beklemishev 1944) as well as protostomes, but so unique that new phyla were erected to accommodate them (i.e., Pogonophora and Vestimentifera) (reviewed by Rouse [2001] and Pleijel et al. [2009]). The conservation of the developmental gene toolkit probably reflects the developmental constraints into the necessary to go step by step through deterministic stereotypic spiral cleavage and larval development (Nielsen 2004). Akin to other polychaetes, the endoderm is necessary not only later for feeding functions, also seen in the metatrochophore larvae prior symbiont infection (Nussbaumer et al. 2006), but also to develop most mesodermal tissue.
Combining the genomic information with tissue-specific transcriptomes allows us to hypothesize that the mesodermal trophosome (Nussbaumer et al. 2006; Bright et al. 2013) is a multifunctional organ with ancestral inherited functions such as hematopoiesis. This trait, we hypothesize belongs to the functional repertoire known from mesodermal chloragogen (extravasal tissue surrounding the gut and blood vessels) derived from the visceral mesoderm in annelids like the trophosome in vestimentiferans (Nussbaumer et al. 2006; Bright et al. 2013). In fact, van der Land and Nørrevang suggested already in 1975, long before the symbionts were detected, that the trophosome in L. luymesi is the nutritive chloragogen tissue (Van der Land 1975). Although overall knowledge is fragmentary, it has been suggested that hematopoiesis in annelids is carried out by visceral as well as somatic mesoderm (Hartenstein 2006; Grigorian and Hartenstein 2013). In various polychaete species, it was localized in particular in the (extravasal) chloragogen tissue, the (intravasal) heart body (Potswald 1969; Friedman and Weiss 1980; Braunbeck and Dales 1984; Fischer 1993), or the somatic peritoneum (Eckelbarger 1976). Our data support the production of hemoglobin in the trophosome. Whether coelomocytes, known to be the immunocompetent cells of eucoelomates (Vetvicka and Sima 2009) including annelids (Dales 1964; Salzet et al. 2006; Cuvillier-Hot et al. 2014), and the hemocytes also develop from trophosomal tissue appears to be likely but remains to be verified.
The trophosome, however, further shows adaptations to new functions such as the well-known intracellular digestion through endosomal-like maturation of symbiosomes, as well as the processing of ammonia and storage of nitrogen waste analogous to the vertebrate liver. Most aquatic invertebrates, including annelids, are virtually ammoniotelic secreting ammonia (Larsen et al. 2011). Surprisingly, Riftia employs an additional ureotelic metabolism similar to terrestrial invertebrates and vertebrates converting toxic ammonia to urea/and or uric acid. Specifically, we found the entire set of genes for a complete urea cycle known to detoxify ammonia in the Riftia genome, with most of them upregulated in the trophosome. Therefore, we hypothesize that the trophosome share similar functions to the liver of vertebrates: instead of secreting nitrogenous waste products through kidneys like in vertebrates or nephridia in annelids, the trophosome was found to store large amounts of uric acid and urea. Uric acid and urea can be utilized as a bioavailable source of N via the catabolic arm of the urea cycle yielding and CO2. Given the lack of urea transporters in the symbiont’s genome, and the presence of active ureases in the trophosome host tissue, this suggests that both the synthesis and breakdown of uric acid and urea is under host control.
What factor(s) might lead to the evolution of this physiological capacity to sequester and metabolize urea and uric acid? It has been shown in other symbioses that the exchange of bioavailable N between symbiotic partners plays an important role in recycling bioavailable N, such as in coral-dinoflagellate symbiosis that show an almost complete retention of bioavailable N (Tanaka et al. 2018). At many deep-sea vents, including those where Riftia thrive, bioavailable N is limited as ammonium and free amino acids are found in pM concentrations (Johnson et al. 1988). Moreover, Riftia are unable to ingest particulate matter so they cannot derive nitrogen from detritus. However, an abundant source of N is nitrate, which is found in deep seawater and can be reduced to ammonium by some microbes (Girguis et al. 2000). Previous studies (Hentschel and Felbeck 1993; Girguis et al. 2000) found that Riftia take up nitrate from their environment, and the symbionts reduce nitrate to ammonium for symbiont and host growth and biosynthesis. However, the Riftia host’s ability to produce urea means that if can sequester bioavailable N that is only available to the host. At first glance, limiting symbiont access to N might be considered a way to control symbiont growth, as seen in cnidarian-Symbiodiniaceae (Xiang et al. 2020). This latter scenario, however, seems unlikely as there is ample bioavailable N (in the form of ammonium) throughout the trophosome in both freshly collected and experimentally tested worms (De Cian et al. 2000; Girguis et al. 2000). Rather, it seems plausible that Riftia’s production of urea allows the host to store and sequester N in a stable, largely nontoxic form. Whether urea is mobilized and provided to the host and symbionts during time of low N availability has yet to be experimentally tested, but this physiological capacity is another example of the remarkable adaptions found within that host, which allow it to modulate the rapid environmental changes found at vents and continue to provide for its own and the symbionts’ metabolic demands.
Although the physiological and evolutionary aspects of tubeworm endosymbiosis have been sufficiently addressed over the past 40 years, the molecular mechanisms regulating host and symbiont interactions in siboglinids are still not fully understood. An immuno-centric view has been explored to explain the maintenance and regulation of the endosymbiont population in the giant tubeworm trophosome (Nyholm et al. 2012; Hinzke et al. 2019). Our results, contrary to the expectations, indicate that genes involved with the innate immune responses are downregulated in the trophosome (e.g., Toll-like receptor/MyD88) or in adult tubeworm tissues (e.g., sushi). These results suggest that the innate immune system plays a more prominent role into the establishment of the symbiosis during the infection in the larval stage, rather than preservation of the mutualism during the juvenile/adult life cycle. The control of the endosymbiont population in the trophosome is mainly achieved by the upregulation of endosomal and lysosomal hydrolases resulting in the active digestion of the endosymbionts (a “mowing” process as described by Hinzke et al. 2019).
The giant tubeworm genome establishes a unique and unprecedent hallmark bridging more than four decades of physiological research in Riftia, whereas it simultaneously provides new insights into the development, whole organism function and evolution of one of the most studied models for metazoan–symbiont interaction. We envisage that the resources generated herein foster many hypothesis-driven research pointing toward a more complete understanding of the genotype/phenotype interface in the Riftia and closely related taxa.
Materials and Methods
A detailed methods section is available in supplementary materials and methods and supplementary figure 61, Supplementary Material online. A brief overview of the bioinformatics pipeline follows below.
Biological Material and Sequencing
Riftia genome DNA was obtained from a piece of vestimentum tissue belonging to single worm collected at the hydrothermal vent site Tica, East Pacific Rise (Alvin dive 4839, 9°50.398′N, 104°17.506′W, 2,514 m depth, 2016) (supplementary figs. 1 and 2, Supplementary Material online). PacBio libraries were generated with Sequel technology using the purified Riftia DNA. Tissue-specific transcriptomes were obtained from two specimens collected at Guaymas Basin, one female from the vent site Rebecca’s Roost (SuBastian dive 231, 27°0.645′N, 111°24.418′W, 2,012 m depth, 2019) and one male from a vent site close to Big Pagoda (SuBastian dive 233, 27°0.823′N, 111°24.663′W, 2,015 m depth, 2019) (supplementary figs. 1 and 2, Supplementary Material online). The eight-stranded paired-end tissue-specific transcriptomes (2×150 pb) were sequenced using Illumina NovaSeq SP technology.
Genome Assembly and Processing
The five Riftia PacBio libraries were mapped against a custom database built with Riftia mitochondrial genome and its complete Endoriftia genome using minimap v2.17-r941 (Li 2018). Genome assembly was performed with canu v1.8 (Koren et al. 2017) with optimized parameters. Genome preprocessing, polishing, haplotig removal, and contamination screening were performed with arrow v2.3.3 (https://github.com/pacificbiosciences/genomicconsensus), purge_dups (https://github.com/dfguan/purge_dups), and blobtools v1.1.1 (Laetsch and Blaxter 2017), respectively. Mitochondrial and endosymbiont genome assemblies were executed with flye v2.5 (Kolmogorov et al. 2019). Annotation of the mitochondrial genome was performed with MITOS2 and GeSeq (Bernt et al. 2013; Tillich et al. 2017).
Transcriptome Assembly and Processing
The removal of adapter sequences and quality filtering of reads from the raw transcriptome databases were performed with bbduk v38.42 (https://sourceforge.net/projects/bbmap/). De novo and genome-guided transcriptome assemblies were performed with transabyss v.2.0.1 and STAR v2.7.1a—Stringtie v2.0.6, respectively (Robertson et al. 2010; Dobin et al. 2013; Kovaka et al. 2019). Endoriftia contamination, if present, was removed from the transcriptomes using BlastN v2.8.1+ (Camacho et al. 2009). A global de novo transcriptome was generated with corset and Lace (https://github.com/Oshlack/Lace) (Davidson and Oshlack 2014).
Gene Prediction and Annotation
The repeat landscape of Riftia genome was identified combining a custom giant tubeworm RepeatModeler v2.0 library followed by the masking of the repetitive elements with RepeatMasker v4.0.9 (Smit AFA, Hubley R, Green P, RepeatMasker Open-4.0). Ab initio gene prediction was performed with Augustus v3.3.3 aided with hint files (Stanke and Morgenstern 2005; Hoff and Stanke 2018). Only gene models with homology, orthology, and gene expression evidence were kept. Protein annotation was performed with Interproscan v5.39-77.0, RNAscan-SE 2.0.5, signalP v5.0b, and pfam_scan.pl (Jones et al. 2014; Lowe and Chan 2016; Almagro Armenteros et al. 2019).
Identification of Gene Toolkits in Riftia
Riftia protein sequences were searched against well-curated catalog of developmental genes, amino and fatty acid biosynthesis, endocytosis-, apoptosis, autophagy-, and immune-related genes using BlastP v2.8.1+ and KEGG Automatic Annotation server (https://www.genome.jp/kegg/kaas/) (Moriya et al. 2007). Additionally, protein domain information was retrieved from pfam_scan.pl and Interpro results. Homology of the identified genes was confirmed through phylogenetic inferences using iqtree v1.6.11 combining ModelFinder, tree search, 1,000 ultra-fast bootstrap, and SH-aLRT test replicates (Nguyen et al. 2015; Kalyaanamoorthy et al. 2017; Hoang et al. 2018). The protein diagrams were drawn using IBS v.1.0.3 software (Liu et al. 2015), and the clustered heatmaps generated with the R package pheatmap (v1.0.12). Quantification of the gene expression levels was performed with kallisto v0.46.1 (Bray et al. 2016).
Orthology, Gene Family Analysis, and Positively Selected Genes
To assess Riftia, Lamellibrachia and Annelida lineage-specific genes, orthology inferences using selected nonbilaterian, deuterostome, lophotrochozoan, ecdysozoans, representatives (N = 36) were performed with Orthofinder v2.3.8 (Emms and Kelly 2019). To identify statistically significant gene family expansions/contractions in Riftia compared with other lophotrochozoans, a second round of orthology was performed using 18 lophotrochozoan representatives and Tribolium castaneum as outgroup. Finally, to identify the gene family core within Annelida, a last instance of orthofinder v2.3.8 was invoked using the C. teleta, H. robusta, L. luymesi, and R. pachyptila. Only the longest isoform for each gene was used in the analysis. Nonsynonymous (Ka) and synonymous (Ks) substitution rates were calculated with the stand-alone version of KaKs_calculator v.2 and HyPhy v. 2.5.15 (Pond et al. 2005; Wang et al. 2010; Zhang, Xiao, et al. 2012). Only single-copy genes (1:1 orthologs) without any inconsistencies between the nucleotide and protein sequences were used in the analyses. Contracted and expanded gene families in the giant tubeworm genome were identified using CAFE v4.2.1 (De Bie et al. 2006; Han et al. 2013) using a calibrated starting tree produced by Phylobayes v4.1b (Lartillot et al. 2013). The contracted/expanded gene families were annotated with Interproscan v5.39-77.0 and the enrichment analysis for GO was performed with topGO v2.36.0 using Fisher’s exact test against the R. pachyptila background (i.e., complete set of Riftia genes) coupled with weight01 algorithm. Rapidly evolving gene families in Riftia were annotated using PANTHER HMM scoring tool v2.2 with PANTHER_hmmscore database v15 (Mi et al. 2017). Protein domain contractions and expansions were found using iterative two-tailed Fisher’s exact (supplementary file 2, Supplementary Material online) test applied to pfam_scan.pl results. The obtained P values were corrected using Benjamini and Hochberg method (Benjamini and Hochberg 1995) and only domains with a significant P value of <0.01 were further investigated.
Hemoglobin Evolution
The predicted Riftia hemoglobin (Hb) protein sequences were interrogated for the presence of the globin domain (PF00042) with hmmalign v3.1b2 (Mistry et al. 2013) and proteins without a hit were excluded from the analyses. Manual inspection and characterization of the signature diagnostic residues/motifs in the hemoglobin chain and linker sequences were performed following previous works (Belato et al. 2019). Phylogenetic analyses were carried out as described in the section “Identification of Gene Toolkits in Riftia.” The resulting trees were midpoint rooted using Figtree (http://tree.bio.ed.ac.uk/software/figtree/). Additionally, to investigate the hemoglobin gene expression across different environmental conditions (sulfur rich, sulfur depleted, and medium), we downloaded six publicly available trophosome transcriptomes from SRA (https://www.ncbi.nlm.nih.gov/sra) (accession nos. SRR8949066–SRR8949071). The transcriptome libraries were preprocessed as described in the section “Transcriptome Assembly and Processing.” Riftia Hb sequence was modeled using the Prime program implemented in the Schrödinger Drug Discovery (v2020.2) software suite. All illustrations of structures were made with PyMol v2.4 (https://pymol.org/2/).
Comparative Tissue-Specific Transcriptome
The Riftia transcriptome libraries were pseudoaligned against the merged filtered AUGUSTUS gene models with kallisto v.0.46.1 (Bray et al. 2016) to collect the gene expression data expressed as TPM counts (transcripts per million). Normalization within and across tissues was independently performed before calculating the tissue specificity tau values (see https://rdrr.io/github/roonysgalbi/tispec/f/vignettes/UserGuide.Rmd). To mitigate possible sex-specific differences in the gene expression levels, tau calculations were performed using only the tubeworm female tissues. The absolutely TSGs (genes expressed only in a single tissue defined by a tau value of 1) were submitted to enrichment analyses for GO with topGO as mentioned in the section “Orthology, Gene Family Analysis, and Positively Selected Genes.”
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Christian Baranyi (University of Vienna) and Jennifer Delaney (Harvard University) for the technical support on the RNA/DNA extraction and purification protocols, respectively. We also thank the Vienna BioCenter Core Facilities and the Genomics Center of the University of Minnesota for the generation of Riftia’s “omics” data. Finally, all authors thank the captains and crews of R/V Atlantis and R/V Falkor and the crews of the submersible Alvin and ROV SuBastian for their support throughout the cruise in 2016 and 2019. A.L.O. thanks Thales Kronenberger (Universität Klinikum Tübingen) for the help with the hemoglobin 3D homology modeling and Yanan Sun for the small collaborative work into the investigation of the Hox complement in Lamellibrachia. A.L.O. also thanks Andrew Calcino and Salvador Espada Hinojosa for their constructive conversations during the development of this work, and finally Bruna Yuri Pinheiro Imai for her continuous support to the first author. This work was funded by the Austrian Science Fund (Förderung der wissenschaftlichen Forschung - FWF) project P 31543 granted to M.B.
Author Contributions
A.L.O., M.B., and P.G. designed the project. A.L.O. generated, implemented, and executed all the bioinformatic pipelines. All data analysis was performed by A.L.O. with input from M.B. and P.G., and J.M. A.L.O. wrote the first complete draft of this manuscript and all authors read, commented on, and approved the final version.
Data Availability
The raw long and short reads used to generate the draft genome and tissue-specific transcriptomes, respectively, are available in the SRA database under the BioProject number PRJNA754493 (supplementary table 12, Supplementary Material online). The assembled draft genome, transcriptomes, ab initio-predicted gene models, and annotation are deposited in Phaidra, a permanent repository at the University of Vienna, under the following online address: https://phaidra.univie.ac.at/detail/o:1220865. Supplementary tables, scripts, AUGUSTUS, and RepeatModeler/Masker files generated in this study are included in the supporting files of this manuscript, Supplementary Material online.
References
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H.. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 37(4):420–423. [DOI] [PubMed] [Google Scholar]
- Arp AJ, Childress JJ.. 1981. Blood function in the hydrothermal vent vestimentiferan tube worm. Science 213(4505):342–344. [DOI] [PubMed] [Google Scholar]
- Arp AJ, Childress JJ.. 1983. Sulfide binding by the blood of the hydrothermal vent tube worm Riftia pachyptila. Science 219(4582):295–297. [DOI] [PubMed] [Google Scholar]
- Arp AJ, Childress JJ, Vetter RD.. 1987. The sulphide-binding protein in the blood of the vestimentiferan tube-worm, Riftia pachyptila, is the extracellular haemoglobin. J Exp Biol. 128(1):139–158. [Google Scholar]
- Bailly X, Jollivet D, Vanin S, Deutsch J, Zal F, Lallier F, Toulmond A.. 2002. Evolution of the sulfide-binding function within the globin multigenic family of the deep-sea hydrothermal vent tubeworm Riftia pachyptila. Mol Biol Evol. 19(9):1421–1433. [DOI] [PubMed] [Google Scholar]
- Beklemishev VN. 1944. Osnovy sravintel’noi anatomii bespozvonochnykh. [Principles of the comparative anatomy of invertebrates]. Mosc Akad Nauk [Internet]. 698p. Available from: https://www.marinespecies.org/aphia.php?p=sourcedetails&id=66718. [Google Scholar]
- Belato FA, Schrago CG, Coates CJ, Halanych KM, Costa-Paiva EM.. 2019. Newly discovered occurrences and gene tree of the extracellular globins and linker chains from the giant hexagonal bilayer hemoglobin in metazoans. Genome Biol Evol. 11(3):597–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 57(1):289–300. [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Bonnivard E, Catrice O, Ravaux J, Brown SC, Higuet D.. 2009. Survey of genome size in 28 hydrothermal vent species covering 10 families. Genome 52(6):524–536. [DOI] [PubMed] [Google Scholar]
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunbeck T, Dales RP.. 1984. The role of the heart-body and of the extravasal tissue in disposal of foreign cells in two polychaete annelids. Tissue Cell. 16(4):557–563. [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 34(5):525–527. [DOI] [PubMed] [Google Scholar]
- Bright M, Eichinger I, von Salvini-Plawen L.. 2013. The metatrochophore of a deep-sea hydrothermal vent vestimentiferan (Polychaeta: Siboglinidae). Org Divers Evol. 13(2):163–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright M, Keckeis H, Fisher CR.. 2000. An autoradiographic examination of carbon fixation, transfer and utilization in the Riftia pachyptila symbiosis. Mar Biol. 136(4):621–632. [Google Scholar]
- Bright M, Lallier F.. 2010. The biology of vestimentiferan tubeworms. In: Gibson RN, Atkinson RJA, Gordon JDM, editors. Oceanography and marine biology: an animal review. Vol. 48. United Kingdom: CRC Press-Taylor & Francis Group. p. 213–265. [Google Scholar]
- Bright M, Sorgo A.. 2005. Ultrastructural reinvestigation of the trophosome in adults of Riftia pachyptila (Annelida, Siboglinidae). Invertebr Biol. 122(4):347–368. [Google Scholar]
- Calcino AD, de Oliveira AL, Simakov O, Schwaha T, Zieger E, Wollesen T, Wanninger A.. 2019. The quagga mussel genome and the evolution of freshwater tolerance. DNA Res. 26(5):411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caullery M. 1914. Sur les Siboglinidae, type nouveau d’invertébrés recueillis par l’expédition du Siboga. Bull Soc Zool Fr. 39:350–353. [Google Scholar]
- Cavanaugh CM, Gardiner SL, Jones ML, Jannasch HW, Waterbury JB.. 1981. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science 213(4505):340–342. [DOI] [PubMed] [Google Scholar]
- Childress JJ, Fisher CR.. 1992. The biology of hydrothermal vent animals: physiology, biochemistry, and autotrophic symbioses. Unkn J. 30:337–441. [Google Scholar]
- Childress JJ, Girguis PR.. 2011. The metabolic demands of endosymbiotic chemoautotrophic metabolism on host physiological capacities. J Exp Biol. 214(Pt 2):312–325. [DOI] [PubMed] [Google Scholar]
- Cho S-J, Vallès Y, Giani VC, Seaver EC, Weisblat DA.. 2010. Evolutionary dynamics of the wnt gene family: a Lophotrochozoan perspective. Mol Biol Evol. 27(7):1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cian M-CD, Bailly X, Morales J, Strub J-M, Dorsselaer AV, Lallier FH.. 2003. Characterization of carbonic anhydrases from Riftia pachyptila, a symbiotic invertebrate from deep-sea hydrothermal vents. Proteins Struct Funct Bioinforma. 51(3):327–339. [DOI] [PubMed] [Google Scholar]
- Cian MD, Regnault M, Lallier FH.. 2000. Nitrogen metabolites and related enzymatic activities in the body fluids and tissues of the hydrothermal vent tubeworm Riftia pachyptila. J Exp Biol. 203(Pt 19):2907–2920. [DOI] [PubMed] [Google Scholar]
- Corliss JB, Dymond J, Gordon LI, Edmond JM, von Herzen RP, Ballard RD, Green K, Williams D, Bainbridge A, Crane K, et al. 1979. Submarine thermal springs on the Galápagos rift. Science 203(4385):1073–1083. [DOI] [PubMed] [Google Scholar]
- Cuvillier-Hot V, Boidin-Wichlacz C, Tasiemski A.. 2014. Polychates as annelid models to study ecoimmunology of marine organisms. J Mar Sci Technol. 22(1):9–14.
- Dales RP. 1964. The coelomocytes of the terebellid polychaete Amphitrite johnstoni. J Cell Sci. s3–105(70):263–279. [Google Scholar]
- Davidson NM, Oshlack A.. 2014. Corset: enabling differential gene expression analysis for de novoassembled transcriptomes. Genome Biol. 15(7):410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie T, Cristianini N, Demuth JP, Hahn MW.. 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22(10):1269–1271. [DOI] [PubMed] [Google Scholar]
- De Cian M-C, Regnault M, Lallier F.. 1997. Nitrogenous metabolites in tissues and circulating fluids of Riftia pachyptila. Cah Biol Mar. [Internet]. 38(2):122. Available from: http://www.vliz.be/en/imis?refid=66292. [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. 2010. The symbiotic habit. Princeton: Princeton University Press. [Google Scholar]
- Duboule D. 2007. The rise and fall of Hox gene clusters. Development 134(14):2549–2560. [DOI] [PubMed] [Google Scholar]
- Eckelbarger KJ. 1976. Larval development and population aspects of the reef-building polychaete Phragmatopoma lapidosa from the east coast of Florida. Bull Mar Sci. 26:117–132. [Google Scholar]
- Ellers J, Kiers ET, Currie CR, McDonald BR, Visser B.. 2012. Ecological interactions drive evolutionary loss of traits. Ecol Lett. 15(10):1071–1082. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Diez C, Miguel V, Mennerich D, Kietzmann T, Sánchez-Pérez P, Cadenas S, Lamas S.. 2015. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 6:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchald K, Rouse G.. 1997. Polychaete systematics: past and present. Zool Scripta. 26(2):71–138. [Google Scholar]
- Felbeck H. 1981. Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila Jones (Vestimentifera). Science 213(4505):336–338. [DOI] [PubMed] [Google Scholar]
- Felbeck H. 1985. CO2 fixation in the hydrothermal vent tube worm Riftia pachyptila (Jones). Physiol Zool. 58(3):272–281. [Google Scholar]
- Felbeck H, Jarchow J.. 1998. Carbon release from purified chemoautotrophic bacterial symbionts of the hydrothermal vent tubeworm Riftia pachyptila. Physiol Zool. 71(3):294–302. [DOI] [PubMed] [Google Scholar]
- Felbeck H, Turner PJ.. 1995. CO2 transport in catheterized hydrothermal vent tubeworms, Riftia pachyptila (vestimentifera). J Exp Zool. 272(2):95–102. [Google Scholar]
- Fernández R, Gabaldón T.. 2020. Gene gain and loss across the metazoan tree of life. Nat Ecol Evol. 4(4):524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiermonte G, Paradies E, Todisco S, Marobbio CMT, Palmieri F.. 2009. A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme A and adenosine 3′,5′-diphosphate in human mitochondria. J Biol Chem. 284(27):18152–18159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E. 1993. The myelo-erythroid nature of the chloragogenous-like tissues of the annelids. Comp Biochem Physiol A Physiol. 106(3):449–453. [Google Scholar]
- Fisher CR, Childress JJ, Arp AJ, Brooks JM, Distel D, Favuzzi JA, Macko SA, Newton A, Powell MA, Somero GN, et al. 1988. Physiology, morphology, and biochemical composition of Riftia pachyptila at Rose Garden in 1985. Deep Sea Res Part Oceanogr Res Pap. 35(10–11):1745–1758. [Google Scholar]
- Flores JF, Fisher CR, Carney SL, Green BN, Freytag JK, Schaeffer SW, Royer WE.. 2005. Sulfide binding is mediated by zinc ions discovered in the crystal structure of a hydrothermal vent tubeworm hemoglobin. Proc Natl Acad Sci U S A. 102(8):2713–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman MM, Weiss L.. 1980. An electron microscopic study of hemoglobin synthesis in the marine annelid, Amphitrite ornata (Polychaeta: Terebellidae). J Morphol. 164(2):121–138. [DOI] [PubMed] [Google Scholar]
- Gaill F, Hunt S.. 1986. Tubes of deep sea hydrothermal vent worms Riftia pachyptila (Vestimentifera) and Alvinella pompejana (Annelida). Mar Ecol Prog Ser. 34:267–274. [Google Scholar]
- Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME.. 2006. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 90(1):5–22. [DOI] [PubMed] [Google Scholar]
- Gardebrecht A, Markert S, Sievert SM, Felbeck H, Thürmer A, Albrecht D, Wollherr A, Kabisch J, Le Bris N, Lehmann R, et al. 2012. Physiological homogeneity among the endosymbionts of Riftia pachyptila and Tevnia jerichonana revealed by proteogenomics. ISME J. 6(4):766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner SL, Jones ML.. 1993. Microscopic anatomy of invertebrates. New York: Wiley-Liss. [Google Scholar]
- Gardiner SL, Jones ML.. 1994. On the significance of larval and juvenile morphology for suggesting phylogenetic relationships of the Vestimentifera. Am Zool. 34(4):513–522. [Google Scholar]
- Gazave E, Lemaître QIB, Balavoine G.. 2017. The Notch pathway in the annelid Platynereis: insights into chaetogenesis and neurogenesis processes. Open Biol. 7(2):160242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdol M, Venier P, Pallavicini A.. 2015. The genome of the Pacific oyster Crassostrea gigas brings new insights on the massive expansion of the C1q gene family in Bivalvia. Dev Comp Immunol. 49(1):59–71. [DOI] [PubMed] [Google Scholar]
- Girguis PR, Childress JJ, Freytag JK, Klose K, Stuber R.. 2002. Effects of metabolite uptake on proton-equivalent elimination by two species of deep-sea vestimentiferan tubeworm, Riftia pachyptila and Lamellibrachia cf luymesi: proton elimination is a necessary adaptation to sulfide-oxidizing chemoautotrophic symbionts. J Exp Biol. 205(Pt 19):3055–3066. [DOI] [PubMed] [Google Scholar]
- Girguis PR, Lee RW, Desaulniers N, Childress JJ, Pospesel M, Felbeck H, Zal F.. 2000. Fate of nitrate acquired by the tubeworm Riftia pachyptila. Appl Environ Microbiol. 66(7):2783–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffredi S, Childress J, Desaulniers N, Lee R, Lallier F, Hammond DO.. 1997. Inorganic carbon acquisition by the hydrothermal vent tubeworm Riftia pachyptila depends upon high external PCO2 and upon proton-equivalent ion transport by the worm. J Exp Biol. 200(Pt 5):883–896. [DOI] [PubMed] [Google Scholar]
- Goffredi SK, Girguis PR, Childress JJ, Desaulniers NT.. 1999. Physiological functioning of carbonic anhydrase in the hydrothermal vent tubeworm Riftia pachyptila. Biol Bull. 196(3):257–264. [DOI] [PubMed] [Google Scholar]
- Govenar B, Fisher CR.. 2007. Experimental evidence of habitat provision by aggregations of Riftia pachyptila at hydrothermal vents on the East Pacific Rise. Mar Ecol. 28(1):3–14. [Google Scholar]
- Govenar B, Le Bris N, Gollner S, Glanville J, Aperghis A, Hourdez S, Fisher C.. 2005. Epifaunal community structure associated with Riftia pachyptila aggregations in chemically different hydrothermal vent habitats. Mar Ecol Prog Ser. 305:67–77. [Google Scholar]
- Grassle JF. 1987. The ecology of deep-sea hydrothermal vent communities. In: Blaxter JHS, Southward AJ, editors. Advances in marine biology. Vol. 23. United States: Academic Press. p. 301–362. [Google Scholar]
- Grigorian M, Hartenstein V.. 2013. Hematopoiesis and hematopoietic organs in arthropods. Dev Genes Evol. 223(1–2):103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijarro-Clarke C, Holland PWH, Paps J.. 2020. Widespread patterns of gene loss in the evolution of the animal kingdom. Nat Ecol Evol. 4(4):519–523. [DOI] [PubMed] [Google Scholar]
- Halanych KM, Feldman RA, Vrijenhoek RC.. 2001. Molecular evidence that Sclerolinum brattstromi is closely related to Vestimentiferans, not to frenulate pogonophorans (Siboglinidae, Annelida). Biol Bull. 201(1):65–75. [DOI] [PubMed] [Google Scholar]
- Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW.. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 30(8):1987–1997. [DOI] [PubMed] [Google Scholar]
- Hand SC. 1987. Trophosome ultrastructure and the characterization of isolated bacteriocytes from invertebrate-sulfur bacteria symbioses. Biol Bull. 173(1):260–276. [DOI] [PubMed] [Google Scholar]
- Hartenstein V. 2006. Blood cells and blood cell development in the animal kingdom. Annu Rev Cell Dev Biol. 22:677–712. [DOI] [PubMed] [Google Scholar]
- Hejnol A, Martín-Durán JM.. 2015. Getting to the bottom of anal evolution. Zool Anz J Comp Zool. 256:61–74. [Google Scholar]
- Hentschel U, Felbeck H.. 1993. Nitrate respiration in the hydrothermal vent tubeworm Riftia pachyptila. Nature 366(6453):338–340. [Google Scholar]
- Hessler RR, Smithey WM, Boudrias MA, Keller CH, Lutz RA, Childress JJ.. 1988. Temporal change in megafauna at the Rose Garden hydrothermal vent (Galapagos Rift; eastern tropical Pacific). Deep Sea Res Part Oceanogr Res Pap. 35(10–11):1681–1709. [Google Scholar]
- Hilário A, Capa M, Dahlgren TG, Halanych KM, Little CTS, Thornhill DJ, Verna C, Glover AG.. 2011. New perspectives on the ecology and evolution of siboglinid tubeworms. PLoS One 6(2):e16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinzke T, Kleiner M, Breusing C, Felbeck H, Häsler R, Sievert SM, Schlüter R, Rosenstiel P, Reusch TBH, Schweder T, et al. 2019. Host-microbe interactions in the chemosynthetic Riftia pachyptila symbiosis. mBio 10(6):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Stanke M.. 2018. Predicting genes in single genomes with AUGUSTUS. Curr Protoc Bioinforma. e57. [DOI] [PubMed] [Google Scholar]
- Holstein TW. 2012. The evolution of the Wnt pathway. Cold Spring Harb Perspect Biol. 4(7):a007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten SM, Violante S, Ventura FV, Wanders RJA.. 2016. The biochemistry and physiology of mitochondrial fatty acid β-oxidation and its genetic disorders. Annu Rev Physiol. 78:23–44. [DOI] [PubMed] [Google Scholar]
- Hunt HL, Metaxas A, Jennings RM, Halanych KM, Mullineaux LS.. 2004. Testing biological control of colonization by vestimentiferan tubeworms at deep-sea hydrothermal vents (East Pacific Rise, 9°50′N). Deep Sea Res Part Oceanogr Res Pap. 51(2):225–234. [Google Scholar]
- Indiveri C, Iacobazzi V, Tonazzi A, Giangregorio N, Infantino V, Convertini P, Console L, Palmieri F.. 2011. The mitochondrial carnitine/acylcarnitine carrier: function, structure and physiopathology. Mol Aspects Med. 32(4–6):223–233. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C.. 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 12(6):393–406. [DOI] [PubMed] [Google Scholar]
- Ip JC-H, Xu T, Sun J, Li R, Chen C, Lan Y, Han Z, Zhang H, Wei J, Wang H, et al. 2021. Host-endosymbiont genome integration in a deep-sea chemosymbiotic clam. Mol Biol Evol. 38(2):502–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings RM, Halanych KM.. 2005. Mitochondrial genomes of Clymenella torquata (Maldanidae) and Riftia pachyptila (Siboglinidae): evidence for conserved gene order in annelida. Mol Biol Evol. 22(2):210–222. [DOI] [PubMed] [Google Scholar]
- Johnson KS, , ChildressJJ, , HesslerRR, , Sakamoto-ArnoldCM, , Beehler CL.. 1988. Chemical and biological interactions in the Rose Garden hydrothermal vent field, Galapagos spreading center. D eep Sea Res Part Oceanogr Res Pap. 35:1723–1744. [Google Scholar]
- Jones ML. 1981. Riftia pachyptila Jones: observations on the Vestimentiferan Worm from the Galápagos Rift. Science 213(4505):333–336. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast DJ, Dominguez R.. 2017. The cytoskeleton–autophagy connection. Curr Biol. 27(8):R318–R326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiers ET, West SA.. 2015. Evolving new organisms via symbiosis. Science 348(6233):392–394. [DOI] [PubMed] [Google Scholar]
- Kirkitadze MD, Barlow PN.. 2001. Structure and flexibility of the multiple domain proteins that regulate complement activation. Immunol Rev. 180:146–161. [DOI] [PubMed] [Google Scholar]
- Klose J, Polz MF, Wagner M, Schimak MP, Gollner S, Bright M.. 2015. Endosymbionts escape dead hydrothermal vent tubeworms to enrich the free-living population. Proc Natl Acad Sci U S A. 112(36):11300–11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA.. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 37(5):540–546. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM.. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M.. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20(1):278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML.. 2017. BlobTools: interrogation of genome assemblies. F1000Res. 6:1287. [Google Scholar]
- Larsen EH, Deaton LE, Onken H, O’Donnell M, Grosell M, Dantzler WH, Weihrauch D.. 2011. Osmoregulation and excretion. Compr Physiol. 4:405–573. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J.. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 62(4):611–615. [DOI] [PubMed] [Google Scholar]
- Le Bris N, Govenar B, Le Gall C, Fisher CR.. 2006. Variability of physico-chemical conditions in 9°50′N EPR diffuse flow vent habitats. Mar Chem. 98(2–4):167–182. [Google Scholar]
- Le Bris N, Rodier P, Sarradin P-M, Le Gall C.. 2006. Is temperature a good proxy for sulfide in hydrothermal vent habitats? Cah Biol Mar. 47:465–470. [Google Scholar]
- Le Bris N, Sarradin P-M, Caprais J-C.. 2003. Contrasted sulphide chemistries in the environment of 13°N EPR vent fauna. Deep Sea Res Oceanogr Res Pap. 50(6):737–747. [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kocot KM, Schander C, Santos SR, Thornhill DJ, Halanych KM.. 2015. Mitogenomics reveals phylogeny and repeated motifs in control regions of the deep-sea family Siboglinidae (Annelida). Mol Phylogenet Evol. 85:221–229. [DOI] [PubMed] [Google Scholar]
- Li Y, Tassia MG, Waits DS, Bogantes VE, David KT, Halanych KM.. 2019. Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi. BMC Biol. 17(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y, et al. 2015. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31(20):3359–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Chan PP.. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y-J, Kanda M, Koyanagi R, Hisata K, Akiyama T, Sakamoto H, Sakamoto T, Satoh N.. 2018. Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nat Ecol Evol. 2(1):141–151. [DOI] [PubMed] [Google Scholar]
- Luther GW, Rozan TF, Taillefert M, Nuzzio DB, Di Meo C, Shank TM, Lutz RA, Cary SC.. 2001. Chemical speciation drives hydrothermal vent ecology. Nature 410(6830):813–816. [DOI] [PubMed] [Google Scholar]
- Lutz RA, Shank TM, Evans R.. 2001. Life After Death in the Deep Sea: following immolation by volcanic eruption, the community around a hydrothermal vent recovers spectacularly. Am Sci. 89(5):422–431. [Google Scholar]
- Lutz RA, Shank TM, Fornari DJ, Haymon RM, Lilley MD, Von Damm KL, Desbruyeres D.. 1994. Rapid growth at deep-sea vents. Nature 371(6499):663–664. [Google Scholar]
- Majd H, King MS, Smith AC, Kunji ERS.. 2018. Pathogenic mutations of the human mitochondrial citrate carrier SLC25A1 lead to impaired citrate export required for lipid, dolichol, ubiquinone and sterol synthesis. Biochim Biophys Acta Bioenerg. 1859(1):1–7. [DOI] [PubMed] [Google Scholar]
- Marsh AG, Mullineaux LS, Young CM, Manahan DT.. 2001. Larval dispersal potential of the tubeworm Riftia pachyptila at deep-sea hydrothermal vents. Nature 411(6833):77–80. [DOI] [PubMed] [Google Scholar]
- Martín-Durán JM, Vellutini BC, Marlétaz F, Cetrangolo V, Cvetesic N, Thiel D, Henriet S, Grau-Bové X, Carrillo-Baltodano AM, Gu W, et al. . 2021. Conservative route to genome compaction in a miniature annelid. Nat Ecol Evol. 5(2):231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. 2012. TGFβ signalling in context. Nat Rev Mol Cell Biol. 13(10):616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAnulty SJ, Nyholm SV.. 2017. The role of hemocytes in the Hawaiian bobtail squid, Euprymna scolopes: a model organism for studying beneficial host–microbe interactions. Front Microbiol. 7:2013. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5216023/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh D. 1997. Molecular evidence that echiurans and pogonophorans are derived annelids. Proc Natl Acad Sci U S A. 94(15):8006–8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD.. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45(D1):D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F, Peterson CH, Mullineaux LS, Fisher CR, Mills SW, Sancho G, Johnson GA, Lenihan HS.. 2002. Predation structures communities at deep-sea hydrothermal vents. Ecol Monogr. 72(3):365–382. [Google Scholar]
- Minic Z, Hervé G.. 2003. Arginine metabolism in the deep sea tube worm Riftia pachyptila and its bacterial endosymbiont. J Biol Chem. 278(42):40527–40533. [DOI] [PubMed] [Google Scholar]
- Minic Z, Simon V, Penverne B, Gaill F, Hervé G.. 2001. Contribution of the bacterial endosymbiont to the biosynthesis of pyrimidine nucleotides in the deep-sea tube worm Riftia pachyptila. J Biol Chem. 276(26):23777–23784. [DOI] [PubMed] [Google Scholar]
- Mistry J, Finn RD, Eddy SR, Bateman A, Punta M.. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41(12):e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M.. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35(Web Server issue):W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin C-H.. 2009. The regulation of TGFβ signal transduction. Development 136(22):3699–3714. [DOI] [PubMed] [Google Scholar]
- Mullineaux LS, Fisher CR, Peterson CH, Schaeffer SW.. 2000. Tubeworm succession at hydrothermal vents: use of biogenic cues to reduce habitat selection error? Oecologia 123(2):275–284. [DOI] [PubMed] [Google Scholar]
- Mullineaux LS, Mills SW, Le Bris N, Beaulieu SE, Sievert SM, Dykman LN.. 2020. Prolonged recovery time after eruptive disturbance of a deep-sea hydrothermal vent community. Proc Biol Sci. 287(1941):20202070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux LS, Peterson CH, Micheli F, Mills SW.. 2003. Successional mechanism varies along a gradient in hydrothermal fluid flux at deep-sea vents. Ecol Monogr. 73(4):523–542. [Google Scholar]
- Nakahama S, Nakagawa T, Kanemori M, Fukumori Y, Sasayama Y.. 2008. Direct evidence that extracellular giant hemoglobin is produced in chloragogen tissues in a beard worm, Oligobrachia mashikoi (Frenulata, Siboglinidae, Annelida). Zoolog Sci. 25(12):1247–1252. [DOI] [PubMed] [Google Scholar]
- Nelson D, Fisher C.. 1995. Chemoautotrophic and methanotrophic endosymbiotic bacteria at deep-sea vents and seeps. In: Karl DM.The microbiology of deep-sea hydrothermal vents. Boca Raton (FL: ): CRC. p. 125–167. [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C. 2012. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 13(12):767–779. [DOI] [PubMed] [Google Scholar]
- Nielsen C. 2004. Trochophora larvae: cell‐lineages, ciliary bands, and body regions. 1. Annelida and mollusca. J Exp Zool B Mol Dev Evol. 302(1):35–68. [DOI] [PubMed] [Google Scholar]
- Nielsen C, Brunet T, Arendt D.. 2018. Evolution of the bilaterian mouth and anus. Nat Ecol Evol. 2(9):1358–1376. [DOI] [PubMed] [Google Scholar]
- Nussbaumer AD, Fisher CR, Bright M.. 2006. Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441(7091):345–348. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Song P, Dang J, Bunce C, Girguis PR.. 2012. Expression and putative function of innate immunity genes under in situ conditions in the symbiotic hydrothermal vent tubeworm Ridgeia piscesae. PLoS One 7(6):e38267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W.. 2005. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 6(12):893–904. [DOI] [PubMed] [Google Scholar]
- Pflugfelder B, Cary SC, Bright M.. 2009. Dynamics of cell proliferation and apoptosis reflect different life strategies in hydrothermal vent and cold seep vestimentiferan tubeworms. Cell Tissue Res. 337(1):149–165. [DOI] [PubMed] [Google Scholar]
- Pila EA, Sullivan JT, Wu XZ, Fang J, Rudko SP, Gordy MA, Hanington PC.. 2016. Haematopoiesis in molluscs: a review of haemocyte development and function in gastropods, cephalopods and bivalves. Dev Comp Immunol. 58:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires-daSilva A, Sommer RJ.. 2003. The evolution of signalling pathways in animal development. Nat Rev Genet. 4(1):39–49. [DOI] [PubMed] [Google Scholar]
- Pleijel F, Dahlgren TG, Rouse GW.. 2009. Progress in systematics: from Siboglinidae to Pogonophora and Vestimentifera and back to Siboglinidae. CR Biol. 332(2–3):140–148. [DOI] [PubMed] [Google Scholar]
- Polzin J, Arevalo P, Nussbaumer T, Polz MF, Bright M.. 2019. Polyclonal symbiont populations in hydrothermal vent tubeworms and the environment. Proc Biol Sci. 286(1896):20181281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV.. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21(5):676–679. [DOI] [PubMed] [Google Scholar]
- Potswald HE. 1969. Cytological observations on the so‐called neoblasts in the serpulid Spirorbis. J Morphol. 128(2):241–259. [Google Scholar]
- Powell MA, Somero GN.. 1986. Adaptations to sulfide by hydrothermal vent animals: sites and mechanisms of detoxification and metabolism. Biol Bull. 171(1):274–290. [Google Scholar]
- Quetin LB, Childress JJ.. 1980. Observations on the swimming activity of two bathypelagic mysid species maintained at high hydrostatic pressures. Deep Sea Res Oceanogr Res Pap. 27(5):383–391. [Google Scholar]
- Rau GH. 1981a. Low 15N/14N in hydrothermal vent animals: ecological implications. Nature 289(5797):484–485. [Google Scholar]
- Rau GH. 1981b. Hydrothermal vent clam and tube worm 13C/12C: further evidence of nonphotosynthetic food sources. Science 213(4505):338–340. [DOI] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, et al. 2010. De novo assembly and analysis of RNA-seq data. Nat Methods. 7(11):909–912. [DOI] [PubMed] [Google Scholar]
- Robidart JC, Bench SR, Feldman RA, Novoradovsky A, Podell SB, Gaasterland T, Allen EE, Felbeck H.. 2008. Metabolic versatility of the Riftia pachyptila endosymbiont revealed through metagenomics. Environ Microbiol. 10(3):727–737. [DOI] [PubMed] [Google Scholar]
- Roeder RG. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 21(9):327–335. [PubMed] [Google Scholar]
- Rouse GW. 2001. A cladistic analysis of Siboglinidae Caullery, 1914 (Polychaeta, Annelida): formerly the phyla Pogonophora and Vestimentifera. Zool J Linn Soc. 132(1):55–80. [Google Scholar]
- Salzet M, Tasiemski A, Cooper E.. 2006. Innate immunity in lophotrochozoans: the annelids. Curr Pharm Des. 12(24):3043–3050. [DOI] [PubMed] [Google Scholar]
- Sanchez S, Hourdez S, Lallier FH.. 2007. Identification of proteins involved in the functioning of Riftia pachyptila symbiosis by subtractive suppression hybridization. BMC Genomics 8:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JF, Zimmer F, Bornberg-Bauer E.. 2016. Mechanisms of transcription factor evolution in Metazoa. Nucleic Acids Res. 44(13):6287–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G, Käck H, Lindqvist Y.. 2000. The manifold of vitamin B6 dependent enzymes. Structure 8(1):R1–R6. [DOI] [PubMed] [Google Scholar]
- Schulze A. 2002. Histological and ultrastructural characterization of the intravasal body in Vestimentifera (Siboglinidae, Polychaeta, Annelida). Cah Biol Mar. 43(3):355–358.
- Shank TM, Fornari DJ, Von Damm KL, Lilley MD, Haymon RM, Lutz RA.. 1998. Temporal and spatial patterns of biological community development at nascent deep-sea hydrothermal vents (9°50′N, East Pacific Rise). Deep Sea Res Part II Top Stud Oceanogr. 45(1–3):465–515. [Google Scholar]
- Shillito B, Ravaux J, Gaill F, Delachambre J, Thiébaut E, Childress JJ.. 1999. Preliminary data on carbon production of deep-sea vent tubeworms. Mar Ecol Prog Ser. 183:275–279. [Google Scholar]
- Shively JM, van Keulen G, Meijer WG.. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu Rev Microbiol. 52:191–230. [DOI] [PubMed] [Google Scholar]
- Simakov O, Kawashima T, Marlétaz F, Jenkins J, Koyanagi R, Mitros T, Hisata K, Bredeson J, Shoguchi E, Gyoja F, et al. 2015. Hemichordate genomes and deuterostome origins. Nature 527(7579):459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simakov O, Marletaz F, Cho S-J, Edsinger-Gonzales E, Havlak P, Hellsten U, Kuo D-H, Larsson T, Lv J, Arendt D, et al. 2013. Insights into bilaterian evolution from three spiralian genomes. Nature 493(7433):526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Southward E. 1993. Pogonophora. In: Harrison FW,, Rice ME.. Microscopic anatomy of invertebrates, onychophora, chilopoda and lesser protostomata. New York: Wiley-Liss. p. 327–369. [Google Scholar]
- Stanke M, Morgenstern B.. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33(Web Server issue):W465–W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart FJ, Cavanaugh CM.. 2006. Symbiosis of thioautotrophic bacteria with Riftia pachyptila. In: Overmann J, editor. Molecular basis of symbiosis. Vol. 41. Berlin/Heidelberg: Springer-Verlag. p. 197–225. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhang Y, Xu T, Yang Z, Mu H, Yanjie Z, Lan Y, Fields CJ, Hui JHL, Zhang W, et al. 2017. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat Ecol Evol. 1(5):121. [DOI] [PubMed] [Google Scholar]
- Sun Y, Sun J, Yang Y, Lan Y, Ip JC-H, Wong WC, Kwan YH, Zhang Y, Han Z, Qiu J-W, et al. 2021. Genomic signatures supporting the symbiosis and formation of chitinous tube in the deep-sea tubeworm Paraescarpia echinospica. Mol Biol Evol. 38(10):4116–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmáry E, Smith JM.. 1995. The major evolutionary transitions. Nature 374(6519):227–232. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Suzuki A, Sakai K.. 2018. The stoichiometry of coral-dinoflagellate symbiosis: carbon and nitrogen cycles are balanced in the recycling and double translocation system. ISME J. 12(3):860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger R, Terwilliger N, Bonaventura C, Bonaventura J, Schabtach E.. 1985. Structural and functional properties of hemoglobin from the vestimentiferan Pogonophora, Lamellibrachia. Biochim Biophys Acta BBA Protein Struct Mol Enzymol. 829(1):27–33. [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpl R, Brown JC.. 1996. Supramolecular assembly of basement membranes. Bioessays 18(2):123–132. [DOI] [PubMed] [Google Scholar]
- Tokuda G, Elbourne LDH, Kinjo Y, Saitoh S, Sabree Z, Hojo M, Yamada A, Hayashi Y, Shigenobu S, Bandi C, et al. 2013. Maintenance of essential amino acid synthesis pathways in the Blattabacterium cuenoti symbiont of a wood-feeding cockroach. Biol Lett. 9(3):20121153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL, Tracey A, Bobes RJ, Fragoso G, Sciutto E, et al. 2013. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 496(7443):57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uda K, Tanaka K, Bailly X, Zal F, Suzuki T.. 2005. Phosphagen kinase of the giant tubeworm Riftia pachyptila: cloning and expression of cytoplasmic and mitochondrial isoforms of taurocyamine kinase. Int J Biol Macromol. 37(1–2):54–60. [DOI] [PubMed] [Google Scholar]
- Van der Land J. 1975. The systematic position of Lamellibrachia (Annelida, Vestimentifera). Z Zool Syst Evol Sonderh. 1975:86–101. [Google Scholar]
- Veaudor T, Cassier-Chauvat C, Chauvat F.. 2019. Genomics of urea transport and catabolism in cyanobacteria: biotechnological implications. Front Microbiol. 10:2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetvicka V, Sima P.. 2009. Origins and functions of annelide immune cells: the concise survey. Invertebr Surviv J. 6:138–143. [Google Scholar]
- Wang S, , ZhangJ, , JiaoW, , LiJ, , XunX, , SunY, , GuoX, , HuanP, , DongB, , Zhang L, et al. . 2017. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat Ecol Evol. 1(5):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Y, Zhang Z, Zhu J, Yu J.. 2010. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 8(1):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Fisher RM, Gardner A, Kiers ET.. 2015. Major evolutionary transitions in individuality. Proc Natl Acad Sci U S A. 112(33):10112–10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt K, White HE, Wang L, Bateman OA, Slingsby C, Orlova EV, Wistow G.. 2006. Lengsin is a survivor of an ancient family of class I glutamine synthetases in eukaryotes that has undergone evolutionary re-engineering for a role in the vertebrate eye lens. Structure 14(12):1823–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Lehnert E, Jinkerson RE, Clowez S, Kim RG, DeNofrio JC, Pringle JR, Grossman AR.. 2020. Symbiont population control by host-symbiont metabolic interaction in Symbiodiniaceae-cnidarian associations. Nat Commun. 11(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sun J, Sun Y, Kwan YH, Wong WC, Yanjie Z, Xu T, Feng D, Zhang Y, Qiu J-W, et al. 2020. Genomic, transcriptomic, and proteomic insights into the symbiosis of deep-sea tubeworm holobionts. ISME J. 14(1):135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal F, Lallier FH, Green BN, Vinogradov SN, Toulmond A.. 1996. The multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila II. Complete polypeptide chain composition investigated by maximum entropy analysis of mass spectra. J Biol Chem. 271(15):8875–8881. [DOI] [PubMed] [Google Scholar]
- Zal F, Leize E, Lallier FH, Toulmond A, Dorsselaer AV, Childress JJ.. 1998. S-Sulfohemoglobin and disulfide exchange: the mechanisms of sulfide binding by Riftia pachyptila hemoglobins. Proc Natl Acad Sci U S A. 95(15):8997–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal F, Suzuki T, Kawasaki Y, Childress JJ, Lallier FH, Toulmond A.. 1997. Primary structure of the common polypeptide chain b from the multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila: an insight on the sulfide binding-site. Proteins 29(4):562–574. [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490(7418):49–54. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xiao J, Wu J, Zhang H, Liu G, Wang X, Dai L.. 2012. ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun. 419(4):779–781. [DOI] [PubMed] [Google Scholar]
- Zhou XZ, Lu KP.. 2001. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107(3):347–359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw long and short reads used to generate the draft genome and tissue-specific transcriptomes, respectively, are available in the SRA database under the BioProject number PRJNA754493 (supplementary table 12, Supplementary Material online). The assembled draft genome, transcriptomes, ab initio-predicted gene models, and annotation are deposited in Phaidra, a permanent repository at the University of Vienna, under the following online address: https://phaidra.univie.ac.at/detail/o:1220865. Supplementary tables, scripts, AUGUSTUS, and RepeatModeler/Masker files generated in this study are included in the supporting files of this manuscript, Supplementary Material online.