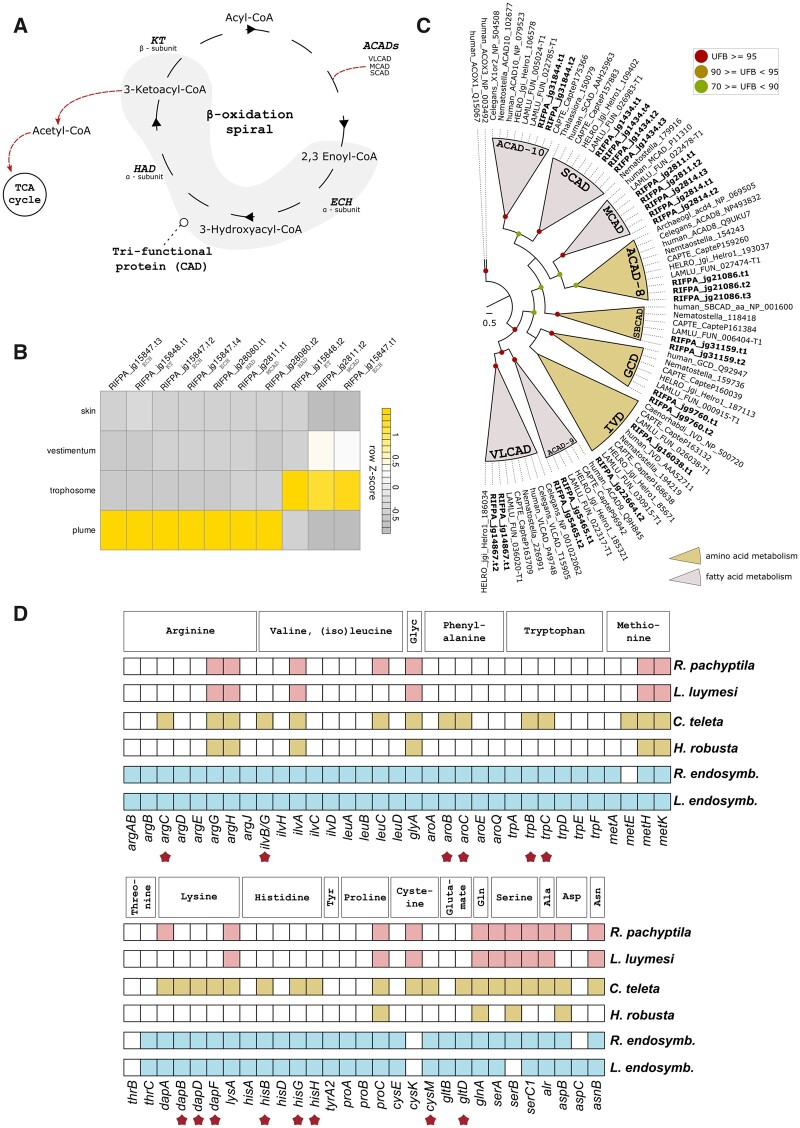
Fig. 4.
Amino acid and fatty acid biosynthesis in Riftia pachyptila. (A) A schematic representation of mitochondrial FAO. The fatty acid degradation is performed in four enzymatic steps involving the membrane bound mitochondrial trifunctional protein and acyl-CoA dehydrogenases. The resulting acetyl-CoA is further oxidized in the TCA cycle. (B) Expression profile of FAO genes. Color coding reflects the expression patterns based on row Z-score calculations. (C) Maximum-likelihood phylogenetic tree inference of the ACAD genes using 1,000 rapid bootstrap replicates. The branch support values are represented by the colored circles in the tree nodes. Accession numbers for NCBI database are displayed after the species names and homologs were retrieved from a previous study (Swigoňová et al. 2009). Capitella, Helobdella, and Lamellibrachia gene identification are derived from the publicly available annotated genomes. (D) Key enzymes related to amino acid biosynthesis identified in Riftia, selected annelids and two tubeworm endosymbiont genomes. The identification of the genes was performed with KEGG and orthology reconfirmed with similarity searches against the ncbi-nr protein database. Riftia and Lamellibrachia lack many amino acid biosynthesis genes indicating nutritional dependence on their endosymbionts. Stars represent genes present in the free-living polychaete Capitella and Endoriftia but absent in Riftia (based on Li et al. [2019] and Tokuda et al. [2013] schemes).