Abstract
The inactivation of antinutritional factors, protease inhibitors within winged bean protein was induced by two respective method treatments. The physical method based on steam vapor that was conducted using an autoclave and chemical method consisting on pH-gradients of buffer solutions prepared at respective acidic pH, neutral pH and alkaline pH ranges. The activity of remaining protease inhibitors of bowman birk inhibitor (BBI), and kunitz-trypsin inhibitor (KTI) after and before treatments was enzymatically confirmed using relevant antagonistic trypsin and combined trypsin-α-chymotrypsin digests. The resulting molecular assembly indicating an interval molecular relaxation range of °0.16 < °DA < °0.2 corresponding to reconformation in protein units with volume-mass changes of −2.17 < ∂v' < +2.17 and with denaturation/unfolding efficiency based on heat capacity ΔCp of 36.36 < DE/UF% < 54.67. These structural changes had a great benefit in determining and producing functional protein hydrolysates.
Keywords: Molecular assembly, Transition energy, Competitive inhibitors, Hydrolysis levels, Denaturation/unfolding, Proteolysates
Graphical abstract
Highlights
-
•
Physical and chemical methods were used in mitigating antinutritional factors.
-
•
The interaction effects of pH, time and temperature have mitigated antinutritional factors.
-
•
Hydrolysis process was conducted to enhance enzyme accessibility to proteins.
1. Introduction
The presence of heat resistant-protease inhibitors along with polyphenols, flavones and phosphatidylcholine in legumes has raised many issues in micronutrient absorption and their stabilities. The structure integrity of those protease inhibitors was distinctive from other storage proteins like soya proteins in terms of structure and function (Roy et al., 2010; Domoney, 1999; Ye and Ng, 2002; Wang and Ng, 2007). One notable feature of the winged bean is the potential for almost all parts of the plant to be eaten, from the seeds to leaves. As a result, extensive researches into their compositions as sources of micro- and macronutrients, as novel food sources for mitigating malnutrition and as neutraceuticals have been conducted. For instance, the dietary guidelines of many countries (USA, 2005) recommend the consumption of 2.5–3.5 cups of pulses per week as part of a healthy diet. Thus, pulses are recognized as functional foods (Maphosa and Jideani, 2017; Muzquiz et al., 2012). Depending on the compounds present in the food matrix, these compounds can acts as anti-nutrients or as bioactive compounds. It is noteworthy that processing of the pulses can decrease or inactivate the content of other non-nutritive compounds such as protease inhibitors, galactosides, lectins, phenols or phytates (Boye et al., 2010). Cooking is more effective in reducing protease inhibitors and lectins, while soaking is more effective in reducing water soluble compounds such as oligosaccharides and other phenolic compounds (Muzquiz et al., 2012). Even though there is not a recommended amount of protease inhibitor consumption. It is important to know that the traditional Japanese diet contains about 420 protease inhibitor units/day. It has been reported as well as that consumption of purified protease inhibitor at 25–800 CIU per day during a period of 12 weeks promoted protective effects against cancer disease development and a dose over 2000 CIU/day did not cause health problem in human (Pedrosa et al., 2021; Muzquiz et al., 2012; Carbonaro, 2011; Singh et al., 2017). It has been also reported that a diet with 1% sodium phytate added can control hypercholestrolaemia and atherosclerosis and it can reduce the risk of colon cancer in the same time it can improve irritable bowel syndrome (Barman et al., 2018; Schlemmer et al., 2009). In addition, some researchers have been reported that iron absorption can be improved when IP6 is below 10 mg/g protein in one serving dose (Fredrikson et al., 2001; Hurrell et al., 1992). Certain amounts of phenolic compounds including tannins are recommended at minimum intake of 300 mg/g where this amount could maintain beneficial effect to human health against maladies and diseases (Oomah et al., 2005). The consumption of legume seeds e.g. winged bean seeds comprising those protease inhibitors before protreating them with heat treatments were capable in disordering enzymatic ingestions, eventually leading to low availability of micronutrients and essential amino-acids (Henley and Kuster, 1994; Deshpande and Damodaran, 1989). The increased demand to these legume seeds made them with comparable nutritive values to soybeans and with high quality of protein and with excellent sources of unsaturated lipids, carbohydrates, minerals and vitamins (Garcia and Palmer, 1980; Prakash, Misra and Misra, 1987; Arun et al., 2003; Arinathan, Mohan and De Britto, 2003; Giami et al., 1992; Cerny et al., 1971). Many of the natural inhibitors with biological activities are present in grain legumes, which might influence directly the physiological changes within the living organisms (Champ, 2002). The impact of these natural compounds has been reported to be positive, negative or both towards the living system, particularly in promoting natural protective effects against numerous maladies and diseases (Champ, 2002; Jamroz and Kubizna, 2008). Most of the interfering compounds are distributed in distinct regions of grain legumes. Among these inhibitory compounds, we have for examples phytic acid, tannic acid, protease inhibitors, and lectin (Liener, 1989; D'Mello, 1995). The chemopreventive roles of these vital substances have been reported to induce many physiological and biological roles including the maintenance and modulating the organs growth, and protection of the body (Brenes, Jansman and Marquardt, 2004; Pusztai, Bardocz and Martín-Cabrejas, 2004). Typically, the natural inhibitors can be categorized into two classes, where the first class is heat susceptible inhibitors like protease inhibitors and lectin. While the second class is heat resistance natural inhibitors such as tannin, and phytic acids (D'Mello, 1995). It was reported that the presence of high amounts of antinutritional factors could induce trans-membrane reaction and other pro-inflammatory responses accomplished by cellular and DNA damages (Garg et al., 1992; Terrill et al., 1994). These antinutritional factors were reducing as well as proteins solubility, proteases activity and their proteolytic activities, proteins-structure stability and release of precursor functional proteins e.g. peptide and/or amino-acids (Liu et al., 1998; Welch, 2002; Raboy, 2009; Kumar et al., 2010). The degradation of structural motifs of antinutritional factors accelerated the course effects of enzymatic hydrolysis and made enzyme- substrate in freely states due to the occurrence of some significant modifications at the spacial structure of proteins (Morrison et al., 2007; Alonso, Orúe and Marzo, 1998; Laskowsky and Kato, 1980; Clemente et al., 2005). In view of these, one of the molecular barriers encountering nutrients bioavailability is the non- stability of protein under gastrointestinal tract conditions including acids, enzymes and bile conditions. This molecular issue is more valid when the functional proteins are containing low levels of covalent chemical lesions such as disulfide groups and susceptible amino acids e.g. essential amino acids constituting of aromatic structure chains. The determination of the molecular statuses of protein polymeric chain is possible using moderate destructive pro-treatment methods based refluxing system such as heat irradiations and/or ions-strength. Under these conditions, the optimization of the thermodynamic parameter changes of enthalpy, entropy, heat capacity and gibbs free energy of transitions using differential scanning calorimetry (DSC) is remaining the pivotal design in determining precise models to those applied thermal and aqueous treatments. DSC is one of the thermal analytical methods used in monitoring the structural changes of proteins in parallel to the effects of mass and heat transfer on structure complexation and aggregation. Thus, DSC is providing real data on the molecular stability and heterogeneity of proteins along with their reconformational levels. Therefore, the main objective of this study was destined in denaturing protease inhibitors and unfolding protein polymeric chain for maximising hydrolysis levels and thus, the release of potential molecular segments with desired health attributes.
2. Materials and methods
2.1. Materials
Winged been seeds (WBS) as raw materials were obtained from Agriculture—Plantation Park of Universiti Putra Malaysia. Reagents and other chemical products used were of HPLC grade and supplied by Merck (Darmstadt, Germany). All buffer systems used including Tris-HCl buffer, acetate buffer, glycine-HCl buffer and phosphate buffer were prepared in the laboratory and stored in cold room at 4 °C. Enzymes employed during hydrolysis (e.g. trypsin, and α-chymotrypsin) were purchased from Sigma (Aldrich, USA) and Merck (Darmstadt, Germany). Antinutritional standards were purchased from Sigma Aldrich, USA; Active Advance and Axon Malaysian Company.
2.2. Degradation and inactivation of anti-nutritional compounds using heat irradiation and heat ionization treatments
The heat irradiation treatment was conducted in preparing triplicate samples using an autoclave apparatus (Saini, 1989). An interval sample mass ranging between 1 and 9 g of the sample was placed into borosilicate bottles, hermitically closed in order to avert any contact of steam vapor leading to protein coagulation. The samples were placed in an autoclave at a depth of 30 cm using support near to water surface. The interval temperature used was ranging between 60 and 121 °C fitted with a maximum pressure of 15 psi (1.03421 Bar) at 121 °C and with interval time ranging between 10 and 60 min. In contrast, the heat ionization treatment was carried out in triplicates using series of buffer solutions including Tris-HCl buffer, acetate buffer, glycine-HCl buffer and phosphate buffer with 0.1 M. 10 ml of those respective buffer solutions that were being adjusted at pH values ranging in between 2.5 and 8 were then used to solubilize 5 g of defatted winged bean seed protein powders (DWBSPs). The prepared mixtures were poured into borosilicate bottles, properly capped, labeled and placed into water baths supported by thermostats and subjected to a constant rate of agitation of 150 rpm for controlling the working temperature at interval ranges of 40–95 °C and ionization time as pro-hydrolysis for 1–6 h. Immediately the resulting samples of both method treatments were collected, covered by aluminum foil and cooled down to 4 °C prior to further analyses.
2.3. Extraction, detection and quantification of antinutritional factors before and after method treatments
The extraction of antinutritional factors from treated and non-treated samples was conducted according to the reported protocol of Anta et al. (2010); Tangendjaja et al. (1980); Verzele and Delahaye (1983) with slight modifications. One g of those treated and non-treated samples was placed in a 50 ml test tube supplemented by 25 ml of extracting sodium acetate solution and segregated from each other with ultra-sonic probe for a period of 5 min. The sample was vigorously extracted using a mechanic shaker for a period of 30 min. The upper phase was isolated after centrifugation at 12,000 rpm for a period of time of 10 min and temperature of 15 °C, for further detection and quantification using high performance liquid chromatography–photo diode array detector (HPLC–PDAD). The extraction and isolation procedures were conducted at least three times under same controlled conditions. The isolated volume of 2 ml of each extract was diluted to 2 ml using sodium acetate M = 0.005, filtered through 0.45 μm nylon filters and injected into HPLC–PDAD (Shimadzu Kyoto, Japan LC 20AT). This analytical machine consisted of a pump system linked to oven model CTO-10ASVP supported by loop injector of 20 μL as a required volume of injection and Photo Diode Array model SPD-M20A PDA detector combined with a DELL Optiplex integrator. The samples were separated in a Prevail C18 column 250 mm × 4.6 mm I.D., particle size 5 μm/l, tech, IL, USA with mobile phase solution of 0.005 M sodium acetate, flowed at a rate of 2 mL/min with an isocratic mode. The peaks were monitored at 254 nm for a run time of 10 min and the concentrations of anti-nutritional compounds were identified based on the compounds standards-retention time over to their range of inhibitory levels of 0.025–0.500 mg/mL. The calibration curves generally are used for fitting the minimum and maximum levels of recovered compounds with their bioactivity levels relevant to some synthetic drugs and/or pro-drugs.
2.4. Optimization of method treatments by response surface methodology fitted by central composite design (RSM—CCD)
The degradation of antinutritional factors using heat irradiation treatment (HIT1) and heat ionization treatment (HIT2) was optimized by response surface methodology-central composite design (RSM—CCD) for establishing a precise regression models between dependent and independent variables. The selected factor ranges of temperature (°C), time (min) and sample mass (g) used for HIT1 method treatment were [60, 121], [10, 60] and [1, 9], respectively, whereas for HIT2 the selected factor ranges of temperature (°C), time (hrs) and pH were [40, 90], [1, 6] and [2.5, 8.5], respectively. These variables were ordered into three levels and coordinated into +/− (α) as axial points, max/min as limits to the interior interval values of less than those of +/− (α) as axial points and the middle point was chosen to be in between the max/min interior interval values. The obtained coordinates of minimum, middle and maximum were facilitated the statistical design of RSM-CCD and its calculation using the second-order polynomial regression model (Myers and Montgomery, 2002; Montgomery, 2001). The errors were minimized by center points-RSM block distributions into 20 runs based center point- sequence reputations. Data treatment and optimization procedures were performed by Minitab v. 14 statistical packages (Minitab Inc., Pennsylvania, USA). Model analysis was done by encrypting p-value of model parameters, coefficient of determination (R2) and p-value of lack-of-fit for providing an indication on the adequacy of the obtained RSM-CCD models (Lee et al., 2000).
2.5. Enzymatic hydrolysis using competitive enzymes-inhibitors assay simulated under gastro- intestinal tract conditions
An in vitro model system was conducted to determine the effects of kunitz trypsin inhibitor (KTI) exerted against trypsin enzyme activity and the effect exerted by bowman birk inhibitor (BBI) against the activity of combined trypsin-α-chymotrypsin enzymes simulated under gastrointestinal tract- conditions. Three model types of samples were chosen including defatted native sample (D-NS), defatted heat irradiated treated sample (D-HIT1) and defatted heat ionized treated sample (D-HIT2). The D-NS was used as control, evidencing the intact and stable form of those inhibitors and their polymorphisms on form heterogeneous complex system with proteins. Under optimum conditions of RSM-CCD, the D-HIT1 and D-HIT2 were then dialyzed for 12 h at 4 °C and hydrolyzed under optimum conditions of the enzymatic hydrolysis process of trypsin and combined trypsin-α-chymotrypsin enzymes. The hydrolysates were prepared by placing an amount of protein mass of 0.3 g in a 30 ml phosphate buffer solution with pH 7. The samples were then subjected to a regular agitation and heated to the enzyme optimum temperature 37 °C at ratio of ∼1:100 (w/w) of enzyme to substrate and remained under heating in a water bath shaker for up 6 h at 150 rpm with an increment time of 1hr between each type of the hydrolysate. The prepared samples were carried out in triplicate using borosilicate bottles and immediately the enzymatic hydrolysis was terminated using a boiling water 100 °C for a period of 10 min. The resulting hydrolysates were then freezed at −18 °C.
2.6. Spectrophotometric assay in determining hydrolysis levels
The degree of hydrolysis was conducted by using the O-phthaldialdehyde (OPA) spectrophotometric assay and the absorbance was recorded in triplicate (Church et al., 1983; Salami et al., 2008; Zarei et al., 2012; Ghanbari et al., 2012; Yea et al., 2014). During enzymatic hydrolysis, cleavage of peptide bonds releases the α-amino groups, which are reacted with OPA in the presence of β-mercaptoethanol forming a complex compound detectable at absorbance of 340 nm.
2.7. Determination of protein denaturation/unfolding and re-conformation energetic parameters using DSC
The determination of the occurring mutation states of those protein prototypes (HIT1, HIT2, DS and NDS) was conducted using differential scanning calorimetry (DSC) Mettler Toledo DSC STARe SW 9.20. In the aluminum pan 5–8 mg of the sample was placed and sealed after complete solubilising of the entire constituent using 15 μL of double distilled (deionised) water. Then the prepared DSC pans were kept for 10 min to ensure a complete folding and unfolding of protein sample via volume expansion and placed into the DSC with reference sample (blank). DSC heating rate was 5 °C/min. The samples were analysed at 27 °C during a scanning period of 20 min and limitation temperature of 95 °C. DSC software was implemented to extract the raw values before undergoing complexation and further integration. The enthalpy (ΔH), entropy (ΔS) and heat capacity (ΔCp) are expressed by the following model equations (1), (2), (3):
| (1) |
| (2) |
| (3) |
where: is the heat flow, is the sample mass, is the transition temperature.
2.8. Model correlation established between DSC parameters linking heat and mass transfer equilibrium point
The model correlation established between DSC parameters of heat flow and mass transfer corresponding to degradation and/or denaturation of the secondary metabolites is determined by the model correlation linking the external forces (1) with those of the internal forces (2) by initiating a novel model correlation at the equilibrium status (3). These models equations (4), (5), (6) were as follows:
| (4) |
| (5) |
| (6) |
where: is the heat flow; is the heat capacity; is the velocity of denaturation/unfolding (DE/UF); k is the thermal conductivity coefficient.
2.9. Denaturation/unfolding efficiency % based energy parameters using DSC thermograms
The determination of the denaturation/unfolding efficiency % was carried out based on the changes in the energy E values expressed in terms ΔH, ΔS, and ΔCp, respectively for respective treatment HIT1 and HIT2 in the presence of non-defatted sample NDS and defatted sample DS according to the following equations (7), (8), (9), (10):
| (7) |
| (8) |
| (9) |
| (10) |
3. Results and discussion
3.1. Model prediction and analysis of the effects of HIT1 and HIT2 on antinutritional factors
Table 1 represents the regression coefficients determined for each response variables. Thus, each response function correspond to respective method HIT1 and HIT2 was adequately displayed in the model parameters Table 1. Results indicated that an adequate denaturation occurred to the backbone molecule levels involving branched and non-branched side chains of carbon, hydrogen and phosphor molecules. Tannic acid (TA) and phytic acid (PA) and under the impacts of HIT1 and combination effects of time and temperature were significantly degraded and disordered by raising progressive the level of heat to the entire chemical bonds accomplished by transfer of moisture, ions and cations, particularly at the tight junction of those antinutritive factors with protein sub-unites. The resulting impacts changed the status of protein sub-unites by inducing a microionization effect in disordering protein unites and chemical lesions. These effects in turn allowed the molecular motions and release of the remaining residual portions of binded water via electrolytes diffusions intermediated by electrons movements and their vibrational status to the surrounding agonistic entities or groups. These results were in agreement with Maga (1982) and Hussain et al. (2011) finding indicating that a slight reduction in the PA content occurred after longer heat processing and germination time. The hydrolytic effects significantly influenced the levels of antinutrients (Kayodé et al., 2006; Feil, 2001). Their effects under various pH values depleted phosphor levels and changed its molecular distribution at highly-attracted unsaturated bonds or charged carbon or hydrogen methylating regions. These effects enhanced the formation of unsaturated chemical bonds through electron and proton-chemical force orientation to more disordered region. These effects were ended by initiating various organic acids with different molecular weights and affinities including adjuvant molecular ligands as conjugating moieties. Based on polynomial regression models displayed in Table 1 some models are reduced from quadratic effects to linear effects for ensuring the accuracy and competence of the model parameters. For instance, PA under the effects of HIT1 showed significant p-value (p < 0.034) by linear effects rather than quadratic effects. The non-significant of PA with HIT2 could be attributed to the stability of PA under the effect of ions-strength. On the other hand, MI as derivative compound of PA and under the effects of HIT1 and HIT2 showed significant p-values (p < 0.001) and (p < 0.005) using linear models rather than quadratic effects. Both treatments indicated that the effects were dependent on time rather than temperature; this is probably attributed to the dephosphorylation of MI along with the interaction time. Moreover, GA which is considered as derivative compound of TA showed significant effect by quadratic effect for HIT1 and linear effect by HIT2. The results demonstrated that both temperature and time are useful for the degradation of GA. TA exhibited significant p-values (p < 0.017) for both method treatments HIT1 and HIT2, where the model become significant when the relative mass was reduced from the model of HIT1, indicating that the dominant factors were temperature and time. For HIT2 all parameters were significant for quadratic model used for the degradation of TA.
Table 1.
Regression coefficients, R2, p-value of lack of fit for the final reduced models.
| Responses |
PA (Y1 in %) |
MI (Y2 in %) |
GA (Y3 in %) |
TA (Y4 in %) |
||||
|---|---|---|---|---|---|---|---|---|
| Coefficient | HIT1 | HIT2 | HIT1 | HIT2 | HIT1 | HIT2 | HIT1 | HIT2 |
| β0 | 4.55945 | 10.4453 | 11.5945* | 41.2699* | −1.36031 | 106.617 | 78.4561* | −42.6272 |
| β1 | −0.08039 | −0.2306 | – | – | −0.05506 | −2.195 | 0.5487 | 2.2003 |
| β2 | 0.27226 | −0.3937 | −0.2625 | −9.9086* | 0.14952 | −16.501 | −0.7871 | 23.1686* |
| β3 | – | −0.2730 | −1.5157 | −4.2864 | 1.48964 | – | – | 1.5023 |
| 0.00109 | 0.0024 | – | – | 0.00142* | 0.011 | −0.0052 | −0.0114 | |
| 0.00100 | −0.0392 | −0.0002 | 1.8621* | 0.00127 | 0.537 | −0.0025 | −2.3548* | |
| – | 0.2817 | −0.0319 | 0.5704* | −0.02639 | – | – | −1.2825* | |
| β12 | −0.00405* | 0.0266 | – | – | −0.00324* | 0.195 | 0.0115* | −0.2777* |
| β13 | – | −0.0297 | – | – | −0.01384* | – | – | 0.0890 |
| β23 | – | −0.2297 | 0.0551* | −0.8845* | – | – | – | 2.4731* |
| R2 | 0.407 | 0.417 | 0.478 | 0.763 | 0.872 | 0.360 | 0.488 | 0.807 |
| p-value | 0.034* | 0.471 | 0.001* | 0.058 (∼*) | 0.001* | 0.060(∼*) | 0.017* | 0.017* |
| Lack of fit (p-value) | 0.655 | 0.000 | 0.049 | 0.634 | 0.065 | 0.053 | 0.622 | 0.000 |
Abbreviation: β0 is a constant, βi, βii and βij are the linear, quadratic and interaction coefficients of the quadratic polynomial equation, respectively. 1: Heating temperature generated during HIT1 and HIT2; 2: Time applied for HIT1 and HIT2; 3: Material mass and pH used for HIT1 and HIT2, respectively. *Significant at p < 0.05.
Table 2 showed that the combined effects of pH and time often in combination with reaction temperature had significant (p < 0.05) impact on the degradation levels of phytic acid (PA), tannic acid (TA), gallic acid (GA), and myoinositol (MI). It means that by increasing further the time other associated factors such as temperature, mass and pH were significantly influenced the stability and structure of those acids as it was indicated by the significance of (p < 0.05) factors interactions. The influence of temperature and time of treatments was found to be almost significant to all responses, justifying the importance of time to macro- and micro-ionization of chemical entities, atom vibrations and covalent and non-covalent chemical lesion relaxations and attractions. The resulting changes in the chemical structure of those acids were then modeled by RSM-CCD by fitting different regression models taking into account the final reduced models as the precise ones. It was pointed out that the efficient parameters in inducing the degradation rates of those substances was the time needed for the bioconversion of chemical energy of the entire molecule structures. Furthermore, the interaction of temperature and time of HIT2 and the interaction of time and mass for HIT1 had no significant effects on MI and TA. Thus, these types of interactions were reduced from the final model equations. Response optimizer indicated the applicability of polynomial regression models at extent level of significance of the response parameters where the exponential fitting to the non-linear model equations of e.g. quadratic ones was remaining the best options in modeling compounds decay and meta-static transitions along with heat and mass transfer. Based on response optimizer parameters, the optimum point reflects the cross-link association between dependent and independent variables accomplished by translational graphics to numerical values. This optimum point for HIT1 had the following parameter levels of temperature, time and mass of 94.75 °C, 10 min and 1 g corresponding to 5.5% GA, 8.43% MI, 5.12% PA and 77.31% TA. In case of HIT2 the obtained optimum point corresponded to temperature, time and pH values of 40 °C, 1hr and pH of 2.5, respectively using glycine HCl resulted in 15.5% GA, 32.6% MI, 2.8% PA and 47.63% TA.
Table 2.
The significance probability (p-value, t-ratio) of regression coefficients in final reduced second-order polynomial models.
| Variables | Significance | Main effects |
Quadratic effects |
Interaction effects |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| x1 | x2 | x3 | x11 | x22 | x33 | x1 x2 | x1 x3 | x2 x3 | ||
| PA%, Y1 (HIT1) | p-value | 0.694 | 0.178 | – | 0.317 | 0.530 | – | 0.034* | – | – |
| t-ratio | −0.402 | 1.419 | – | 1.037 | 0.643 | – | −2.249 | – | – | |
| PA%, Y1 (HIT2) | p-value | 0.535 | 0.899 | 0.921 | 0.329 | 0.872 | 0.117 | 0.420 | 0.287 | 0.405 |
| t-ratio | −0.642 | −0.130 | −0.102 | 1.026 | −0.166 | 1.718 | 0.840 | −1.125 | −0.870 | |
| MI%, Y2 (HIT1) | p-value | – | 0.112 | 0.125 | – | 0.927 | 0.668 | – | – | 0.003* |
| t-ratio | – | −1.697 | −1.633 | – | −0.093 | −0.439 | – | – | 3.514 | |
| MI%, Y2 (HIT2) | p-value | – | 0.016* | 0.216 | – | 0.000* | 0.049* | – | – | 0.058*(∼) |
| t-ratio | – | −2.747 | −1.296 | – | 4.890 | 2.157 | – | – | −2.066 | |
| GA%, Y3 (HIT1) | p-value | 0.554 | 0.100 | 0.013* | 0.010* | 0.089 | 0.341 | 0.001* | 0.013* | – |
| t-ratio | −0.611 | 1.794 | 2.959 | 3.110 | 1.866 | −0.994 | −4.331 | −2.963 | – | |
| GA%, Y3 (HIT2) | p-value | 0.072 | 0.271 | – | 0.067 | 0.306 | – | 0.027* | – | – |
| t-ratio | −2.013 | −1.166 | – | 2.054 | 1.079 | – | 2.584 | – | – | |
| TA%, Y4 (HIT1) | p-value | 0.286 | 0.120 | – | 0.063 | 0.522 | – | 0.017* | – | – |
| t-ratio | 1.109 | −1.657 | – | −2.024 | −0.657 | – | 2.700 | – | – | |
| TA%, Y4 (HIT2) | p-value | 0.092 | 0.043* | 0.868 | 0.172 | 0.013* | 0.039* | 0.024* | 0.329 | 0.017* |
| t-ratio | 1.864 | 2.322 | 0.171 | −1.473 | −3.043 | −2.379 | −2.666 | 1.026 | 2.849 | |
Abbreviation:HIT1, heat irradiation treatment; HIT2, heat ionization treatment; x1, x2, and x3 are temperature (°C), time (min), and mass (g) for HIT1, respectively; x1, x2, and x3 are temperature (°C), time (hrs), and pH for HIT2, respectively. *Significant at p < 0.05.
3.2. Response surface plots in determining HIT1 and HIT2 treatment effects
Thermodynamically, the heat is diffused within the food materials by conduction, convection and radiation. At this state, the material mass tends to reach its equilibrium status either by an endo-energy or via an exo-energy heat flow statuses. The significant interaction effects of HIT1 and HIT2 factors on the degradation levels of PA, TA, MI and GA are important in establishing some model correlations between heat and mass transfer in relation to molecules velocity and conductivity coefficient of bioconverted molecules. The results of speeding-up the course effect of HIT2 with pH values is becoming more significant in releasing further freely acids that were usually accomplished by secondary hydrolysis effects. Fig. 1 shows the response-surface plots resulting by both method treatments HIT1 and HIT2 on the degradation levels of TA and PA, respectively. Their decay parameters seemed to steadily reach their maximum degradation level as a function of time and pH for HIT2 (Fig. 1 (f)) and as a function of temperature and time for HIT1 (Fig. 1 (h)), giving an indication on the occurring of a non-equilibrium status in between structure and regenerated chemical energy.
Fig. 1.
Showing response surface plots of degraded levels of MI (a) and (b), PA (h), TA (c), (e) and (f) and GA (d) and (g) based temperature, time in hrs and pH as interaction effects for HIT2 and temperature, time in min and mass as interaction effects for HIT1.
The surface-plots displaying the interaction effects between temperature and pH were neglected due to the least significant effect obtained by those method treatments for PA and TA. The surface plot results showed significant impacts on molecules bioconversion as a function of pH and time for MI and TA as shown in Fig. 1 (b) and (f). Thus, in the selected ranges of pH and time, low hydrolysis time and low pH individually were insufficient for disordering the unsaturated chemical bonds, and therefore, extent release of acid fractions. The heating temperature had a least effect on the hydrolysis and thus on the bioconversion of TA and PA. In the same time it was considered the dominating factor for HIT1. The combination effect of temperature and time of HIT1 significantly reduced the content of PA (Fig. 1 (h)) as compared to the least effects induced by HIT2. Thus, they were reduced in the final models of HIT2 (Table 1). These results indicated that pH and time were the dominating factors to fractional hydrolysis resulting in progressive decay of antinutritional factors, their structure re-conformation, and isomerization. It was depicted out of the surface plot results that the obtained yields of MI (Fig. 1 (b)) and TA (Fig. 1 (f)) were mainly depend on low pH range and reactive time used for the hydrolysis process. Mirhosseini, Tan, Taherian and Boo (2009) stated that the significant interaction effects resulted in a well-defined convex response surfaces that matched good RSM model parameters. The combined effects of pH and time of hydrolysis indicated as well as that sum of the areas of degraded TA were increased when the pH of buffer was lowered down to 2.5 for a minimum period of 1 h. Thus, the structure of MI and TA became unstable at these conditions of pH as compared to the significant enhancement in the molecules resistance at the neutral pH range.
3.3. The effects of HIT1 and HIT2 method treatments on protease inhibitors
The obtained results of the final reduced models for bowman birk inhibitor (BBI), kunitz trypsin inhibitor (KTI) and lectin were adequately displayed in Table 3. There were no significance in the lack of fit and there were adequate values of coefficients of determinations R2 ranging from 0.607 to 0.963. The R2 values indicating good empirical model fits in between data treatments (Lee et al., 2006). The inactivation and denaturation of those inhibitors were fitted on form polynomial regression models. The linear and quadratic effects of KTI model parameters were significant for HIT1 with a p-value of less than 0.023, whereas the linear form was found to be only significant to HIT2 with a p-value of less than 0.022. The reduction in the second order model equation of KTI to become significant was attributed to its considerable recovery levels as compared to BBI. The high recovery levels of KTI as compared to BBI and lectin were attributed to molecular structural disintegration and their copolymer unite segregations. The high complexity levels of KTI and BBI were attributed to their copolymerizations with protein sub- unites rather than lectin that was found to be tagged more with polyphenol compounds e.g. PA and TA moieties. HIT2 revealed a second order model with significant effect to BBI and first order model with significant effect to HIT1. Their p-values were less than 0.002 for HIT1 and were less than 0.026 to HIT2. The degradation of lectin exhibited significant effects to HIT1 and HIT2 on form linear and quadratic forms with p-values of 0.005 and 0.028, respectively (Table 3). Lack of fits were almost found to be not significant for all reduced models confirming the adequacy of RSM-models in fitting reduced results established in between dependent and independent variables.
Table 3.
Regression coefficients, R2, p-value of lack of fit for the final reduced models.
| Responses |
KTI (Y1, %) |
Lectin (Y2, %) |
BBI (Y3, %) |
|||
|---|---|---|---|---|---|---|
| Coefficient | HIT1 | HIT2 | HIT1 | HIT2 | HIT1 | HIT2 |
| β0 | 10.2620 | 6.6139* | 17.1457* | 0.805433 | −7.91743* | 0.02922 |
| β1 | −0.1757 | – | −0.1099 | −0.025771 | 0.10420* | −0.06021 |
| β2 | 0.0381* | −1.2093* | −0.5440* | −0.752982 | – | 3.38440 |
| β3 | 0.5045* | −0.7258 | 0.1543 | 0.727783 | 2.64682* | −1.20048 |
| 0.0004 | – | 0.0003 | 0.000225 | – | −0.00024 | |
| 0.0005 | – | 0.0027* | −0.010021 | – | −0.07964 | |
| 0.0376 | – | −0.0890* | 0.081745* | – | −0.09475 | |
| β12 | 0.0020 | – | 0.0023* | 0.011974 | – | −0.03554 |
| β13 | 0.0142 | – | −0.0025 | −0.011471* | −0.02639* | 0.04163* |
| β23 | −0.05585* | 0.2469* | 0.0290* | – | – | – |
| R2 | 0.664 | 0.368 | 0.820 | 0.963 | 0.607 | 0.687 |
| p-value | 0.023 | 0.022 | 0.005 | 0.028 | 0.002 | 0.026 |
| Lack of fit (p-value) | 0.003 | 0.165 | 0.589 | 0.567 | 0.236 | 0.557 |
Abbreviation: β0 is a constant, βi, βii and βij are the linear, quadratic and interaction coefficients of the quadratic polynomial equation, respectively. 1: Heating temperature generated via heat irradiation treatment or that generated by heat ionization treatment; 2: Time applied to both treatments; 3: Mass used to heat irradiation treatment or the pH of buffer used to heat ionization treatment.*Significant at p < 0.05.
The interaction effects displayed in Table 4 indicated that the material mass and time exhibited significant effect on KTI using HIT1, while pH and time of HIT2 revealed significant effects. The degradation of BBI was significant for HIT1 by the interaction effect of temperature and material mass and by the interaction effects between temperature and pH for HIT2. Lectin was significantly degraded by HIT1 under the interaction parameters of material mass and time and by the effects of temperature and time of HIT2 (Table 4). The microionization altered the existing chemical lesions by restructuring other anionic, cationic, hydrogenic and hydrophobic links existed in between protease inhibitors, protein subunits and minimal traces of bioconverted antinutrients.
Table 4.
The significance probability (p-value, t-ratio) of regression coefficients in final reduced second-order polynomial models.
| Significance | Main effects |
Quadratic effects |
Interaction effects |
||||||
|---|---|---|---|---|---|---|---|---|---|
| x1 | x2 | x3 | x11 | x22 | x33 | x1x2 | x1x3 | x2x3 | |
| HIT1 (KTI, %) p-value | 0.495 | 0.878a | 0.742a | 0.764 | 0.802 | 0.618 | 0.354 | 0.295 | 0.005a |
| HIT1 (KTI, %) t-ratio | −0.709 | 0.157 | 0.339 | 0.308 | 0.258 | 0.515 | 0.971 | 1.104 | −3.564 |
| HIT2 (KTI, %) p-value | – | 0.043a | 0.059 | – | – | – | – | – | 0.022a |
| HIT2 (KTI, %) t-ratio | – | −2.198 | −2.032 | – | – | – | – | – | 2.544 |
| HIT1 (Lectin, %) p-value | 0.340 | 0.000a | 0.820 | 0.631 | 0.008a | 0.020a | 0.032a | 0.671 | 0.002a |
| HIT1 (Lectin, %) t-ratio | −1.002 | −5.074 | 0.234 | 0.496 | 3.293 | −2.756 | 2.496 | −0.437 | 4.187 |
| HIT2 (Lectin, %) p-value | 0.700 | 0.148 | 0.139 | 0.610 | 0.820 | 0.019a | 0.062 | 0.036a | – |
| HIT2 (Lectin, %) t-ratio | −0.395 | −1.555 | 1.597 | 0.525 | −0.234 | 2.744 | 2.079 | −2.390 | – |
| HIT1 (BBI, %) p-value | 0.015a | – | 0.001a | – | – | – | – | 0.002a | – |
| HIT1 (BBI, %) t-ratio | 2.725 | – | 4.023 | – | – | – | – | −3.673 | – |
| HIT2 (BBI, %) p-value | 0.785 | 0.058 | 0.442 | 0.866 | 0.586 | 0.357 | 0.089 | 0.024a | – |
| HIT2 (BBI, %) t-ratio | −0.279 | 2.115 | −0.797 | −0.172 | −0.562 | −0.962 | −1.868 | 2.625 | – |
aSignificant at p < 0.05.
3.4. Response surface plots in displaying changes in protease inhibitor via HIT1 and HIT2
The changes in BBI, KTI and lectin under the effects of temperature, time, pH and material mass of respective HIT1 and HIT2 using response surface plots are shown in Fig. 2. HIT1 response parameters linking changes in KTI as a function of time and mass and changes in KTI as a function of time and pH of HIT2 displayed well- hyper/hypo-convex surface plots (Fig. 2 (a) and (e)). The occurring decay into KTI was found to be at low interaction time and at low level of relative mass, whereas the decay in KTI by HIT2 was reached its maximum values at extent treatment time correspond to a low pH value. Response surface plot gave a good indication on the decay in the KTI mass and its stability levels when the exerting factors tend to be in equilibrium status relevant to ½ course of time and ½ level of mass used. In the same time the decay in KTI observed for HIT2 took place at optimum range where the stability in KTI levels was noted to be due to the neutral pH range. These effects render the equilibrium status and strength of molecule disruption at half time and at 4 to 5 as pH values.
Fig. 2.
Response surface plots of HIT1 and HIT2 demonstrating the changes in the degradation levels of kunitz trypsin inhibitor KTI as a function of time and mass (a) and as a function of time and pH (e). Changes in the degradation level of lectin as a function of temperature and time (b), as a function of temperature and pH (f) and as a function of time and material mass (c). Changes in the degradation level of bowman birk inhibitor BBI as a function of temperature and mass (d) and as a function of temperature and pH (g).
The changes in the response surface plots of lectin as a function of time and temperature for HIT1 and as a function of temperature and pH for HIT2 were distinctive to both method treatments. The micro-ionization induced by HIT1 displayed a hypo-convex plot that was attributed to the occurring of copolymerization statuses associated to extent release of binded water and electrolytes involving molecule lesions as shown in Fig. 2 (c). It displayed a significant reduction by the effects of time and relative mass levels with adequate reduction in the surface-interface tensions. The effect of macro-ionization in turn induced by buffer of HIT2 did not behave in a manner that gave a hypo-convex plot, evidencing the partial fractionations and denaturation of lectin under pH gradients as shown in Fig. 2 (f). The decay in BBI values was found in a well hypo-convex plot under the interactions factors of temperature and relative mass of HIT1 and in a well hyper-convex plot under the interactions factors of temperature and pH of HIT2 as displayed in Fig. 2 (d) and (g). BBI revealed an isothermal decay region at high temperature levels correspond to low relative mass over 5 g and an isothermal stability region ranging in between 80 and 100 °C as shown in Fig. 2 (d). The extent decay correspond to pH gradient of BBI was at low values of pH of 2.5 and was at adequate temperature levels ranging between 40 and 60 °C as shown in Fig. 2 (g). The well displayed hyper-convex plot to BBI was attributed to its double headed structure motifs. The Bowman Birk Inhibitor BBI has a special molecular structure and its structure is quite similar to allosteric enzyme, where the structure of BBI contains two loops comprising a number of amino acids some involved in the active site of protease inhibitor and some exposed at the interface that are necessary for the stability of the enzymes. BBI influence the reaction via a competitive interaction made by the active site of both headed groups to substrate and this type of protease inhibitor is more flexible with high activation energy as the movement of electron helps in increasing the velocity of enzymes and therefore its interaction mechanism. These changes were due to the effect of pH and ion molecular motions induced further by temperature for triggering mesosphere polymorphic forms like agglomerates. Results showed that the optimum conditions for HIT1 were 75.19 °C, 27.2 min and 1 g leading to KTI, lectin and BBI recovery levels of 4.8%, 2.97%, and 0.58%, respectively. HIT2 revealed optimum conditions of 40 °C, 1.5 h (90 min) and pH of 2.5 of glycine HCl. Their resulting chemical features correspond to this optimum point were 3.9% (KTI), 0.88% (lectin) and 0.57% (BBI).
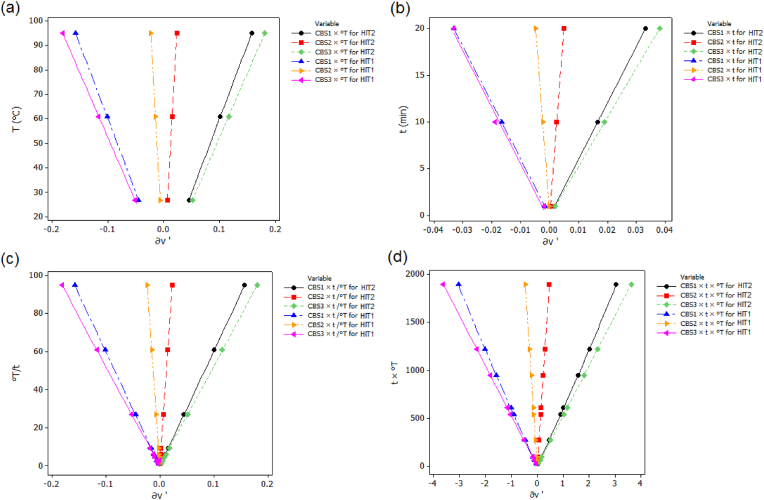
The changes in the expansion volume of protein polymeric chains under micro- and macro-ionization were assigned based on the slight effects of temperature and heating rate (°T/t) as shown in Fig. 3 (a) and (c) with significant changes as a function of heat capacity (t x°T) as shown in Fig. 3 (d) and with least effects induced by time (t) as shown in Fig. 3 (b). The thermal accumulation effect of heat at chemical lesion levels accelerated solute-mass transfer by increasing heat capacity of donating protons and losing electrons. Thus, disordering molecules and their deviation angles, by varying protein surface tensions and their affinity levels. HIT2 and HIT1 increased heat capacity in a range of −4 to +4 °DA as the highest disordering in spacial structure and molecule statuses (Fig. 3 (d)). The heating rates showed optimum °DA ranging between −0.2 and +0.2 °DA in comparison to molecular interval relevant to the impacts of temperature and time as shown in Fig. 3 (a) and (b). Changes in the °DA via attractive and repulsive forces of covalent and non-covalent chemical lesions was attributed to the effects of micro-ionization particularly in inducing reconformation to charged adjacent peptidic chains and due to the cofactor attachments involving glucosidic and phenyl-amine moieties as reducing thiol molecules. These results could be justified as well as by the muta-transitions and instability in the structure of protein accomplished either by the formation of agglomerating regions or glassy states under variable effects of HIT1 and HIT2, respectively. For this reason DSC was conducted to clarify the underlying mechanism of denaturation/unfolding induced by both method treatments HIT1 and HIT2, respectively. DSC procedure was carried out in the presence of two control samples one was defatted from lipids and the other one was remained with lipid bound proteins. In this state the antinutritional factors were degraded within the protein samples using same method treatments of HIT1 and HIT2, respectively. The resulting denaturation/unfolding levels of model equations (7), (8), (9), (10) were determined at the DSC optimum point using RSM response optimizer. Whilst, the in vitro digestibility or hydrolysis mechanism was carried out using optimum condition determined for degradation of protease inhibitors because we are interested in knowing the activity of the enzymes trypsin, trypsin-alpha-chymotrypsin against the remaining levels of protease inhibitors of kunitz-trypsin inhibitor KTI and bowman birk inhibitor BBI. By the way the optimum conditions for protease inhibitors are almost cover the optimum range of protein denaturation/unfolding obtained by DSC.
Fig. 3.
Expansion volume changes due to disordering in chemical bonding statuses as a function of temperature (°c) (a), time (b), °T/t (c) and t × temperature (d).
3.5. In vitro models based region-specificity and entio-selectivity modes of competitive protease inhibitors in determining hydrolysis levels
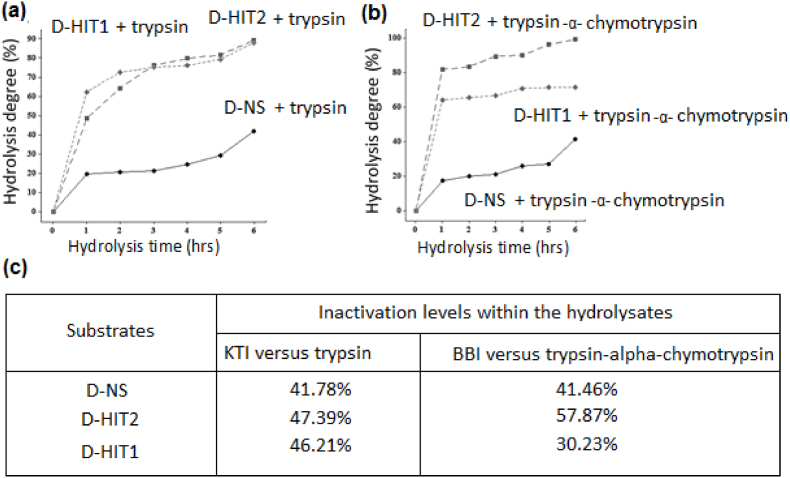
The results of protein hydrolysates using trypsin and combined effects of trypsin-α-chymotrypsin enzymes in the presence of three model types of defatted samples of D-NS, D-HIT2 and D-HIT1 are shown in Fig. 4 (a), (b), (c), and (d). Fig. 4 (a) shows the levels of hydrolysis obtained by trypsin enzyme and trypsin-α-chymotrypsin for a period of time of 6 h. The termination of enzymatic reaction chains of those enzymes was done progressively by assessing the compatibility effects of those enzymes to KTI and BBI molecules as competent inhibitors. As results of their effects on the course of hydrolysis levels assigned to treated samples containing denatured/unfolded competitive inhibitors were in the following orders D-HIT2 (89.17%) > D-HIT1 (87.99%) > D-NS (41.78%). A high level of hydrolysis obtained by D-HIT2 and was attributed to the low level of molecule adherences by decreasing surface and interface tension in between proteins and inhibitors. These results suggested that the structure linearity of remained KTI polymorphic forms were encrypted in crystals aggregation and their deposition in sealing the active site of trypsin enzyme. The results of the hydrolysis obtained by D-HIT1 were closed to those obtained for D-HIT2, reflecting the capability of high temperature-short time HTST in denaturing those effective inhibitors. The D-NS sample exhibited least ingestion levels as compared to D-HIT2 and D-HIT1 samples. These results were due to the presence of considerable peptidic regions involving hydrophilic and hydrophobic regions having similar functions and properties to fibrinogen and fibrolectin. These functional properties led in increasing protein contact surfaces and enzyme accessibility to substrates particularly in traversing distinct conformation levels within the unfolded protein unites. This mechanism effect was the major path in enzymes copolymerizations and their activation energy, renovation and partition. Moreover, the presence of repeated nucleophilic and nucleo-sulfur amino groups could partially inactivate protease inhibitors and therefore, reducing proteases-cleaving lops affinity indices. Thus, the deactivation levels of KTI using D-HIT2 and D-HIT1 ameliorated the hydrolysis levels to 47.39% and 46.21%, respectively although to the remaining minimal traces of protease inhibitors resulting by irradiation and ionization effects.
Fig. 4.
The hydrolysis levels of protein hydrolysates produced by trypsin and trypsin-α-chymotrypsin in competitive hydrolysis with KTI and BBI proteases inhibitors.
This was agreed well with the basic phenomenon of the hydrolysis, where the combination effects of two types of enzymes in the same physiologic working conditions similar to the working mechanism of trypsin and α-chymotrypsin might enhance further the fragmentation mechanism of proteins (Rokka et al., 1997; Saiga et al., 2006; Sauveur et al., 2008). D-HIT2 showed the highest level of hydrolysis of 99.33% by the combined effects of trypsin-α-chymotrypsin followed by D-HIT1 with hydrolysis levels of 71.69% and lastly by D-NS with hydrolysis level of 41.46%. Low level of hydrolysis was recorded to D-NS due to the presence of minimal traces of BBI by exerting feed-back inhibitory effects. D-HIT2 led to a significant mitigation of BBI where the deactivation levels of BBI was 57.87%, whereas D-HIT1 was deactivated BBI only by 30.23%. These results were justified by the compaction of molecules leading in hiding targeted amino acids and due to their least number of repetitions in the targeted protein chains versus enzymes. This in vitro assay has advanced our understanding to the mechanism effect of enzymes, their catalytic efficiency toward other proteases and inhibitors (Mosolov and Valueva, 2005; Qi, Song and Chi, 2005; Clawson, 1996).
3.6. Denaturation/unfolding based HIT1 and HIT2 effects using DSC quantum energy- changes
In the physical and chemical properties of solid, semi- solid and soft materials many of the biochemical reaction would take place after exerting potential forces to the entire constituents leading eventually to atoms disordering states, bioconverted energy, electrolyte diffusion, mass transfer, thermal conductivity, chemical motions, internal friction and mechanical shear. Treating one of these mechanism effects necessitate the interplay of both molecular structure and thermodynamic aspects of food materials presented either on form solid or on form humid states in expressing such mechanism effects of crystallization and copolymerization. These effects in turn affected molecules- contact exposure and their surface- interface tension by rendering the molecular assembly either in a simple matrix network or on form complicated network structure e.g. copolymer conjugates. Enthalpy based model equation (1) in a DSC thermogram as shown in Fig. 5 was the integral part of heat flow as a function of temperature of transition relevant to material mass. This thermodynamic parameter could be changed based on the heating rates and/or heating capacity of structured biomaterials. For instance, ΔH was used in estimating the crystallization of lipids and other additives such as emulsifiers and contaminant lard in lipids moieties (Saadi et al., 2011, 2012a, 2012b; Galeb et al., 2012; Dahimi et al., 2014). ΔH was encrypted in gibbs free energy models linking entropy ΔS values in regenerating mathematical models suitable in fitting biomaterials status changes e.g. flexibility of genius lipopeptides and biopeptides structure motifs destined in scaffolding cell impurities and other diseases growth factors in vivo (Saadi et al., 2015; Saadi et al., 2018). The differences in the ΔH values indicated that the impact of HIT2 was higher than HIT1. These results could be justified by extent energy transmission and release in triggering significant disorders to protein polymeric chains under DSC circumstances and material statuses e.g. mass used, applied heating levels, time, pH levels, polymerization/copolymerization in the presence of minimal traces of antinutrient factors like protease inhibitors, polyphenols and lectin. The obtained model parameters expressed in terms of ΔH for HIT1 and HIT2 are shown in Table 5. Results exhibited significant p-values of 0.018, R2 of 0.618 (61.8%), and insignificant p-value for lack of fit of 0.064 for HIT1, whereas HIT2 regression parameters determined in terms of ΔH values were 0.953 (95.3%), 0.000, and 0.162 for R2, p-value, and p-value for lack of fit, respectively. HIT2 showed excellent coefficient of determination between dependent and independent variables as compared to the moderate R2 exhibited by HIT1. The theoretical concepts of protein conformation and re-conformation are important in understanding the energy behind the stability of protein, their denaturation/unfolding. In this study the entropy ΔS based model equation (2) as one of the quantitative aspects of thermal energy was used in assigning molecules disordered levels. These disordering effects in molecules from position to another position were tentative in returning back to equilibrium status unless if there was a re-shifting in the equilibrium point based on the cumulative effects of energy as shown in Fig. 5 (a) and (b). These effects were more valid when the initial state could be folded via reversible energy of transition leading to improper arrangement to molecules and consistency in the network structure of proteins. The ΔS values using DSC thermograms were distinguished in between the base lines of the native sample and that of the end of the denaturation lines. The model parameters of ΔS were displayed in Table 5 relevant to protein stability and denaturation/unfolding ranges generated by DSC under the influence of method treatments of HIT1 and HIT2. HIT1 revealed the following model parameters of R2 of 0.478 (47.8%), p-value of 0.026 and p-value of lack of fit of 0.621, respectively, whereas HIT2 exhibited R2 of 0.907 (90.7%), significance in p-value of 0.000 and non-significance of p-value of lack of fit of 0.284, respectively. It was concluded that RSM-CCD was successfully determined the impact of processing factors, eventually leading to appropriate model correlations in between response variables and methods conditions. Thus, its suitability and applicability to further biological matters including carbohydrates and lipids … etc.
Fig. 5.
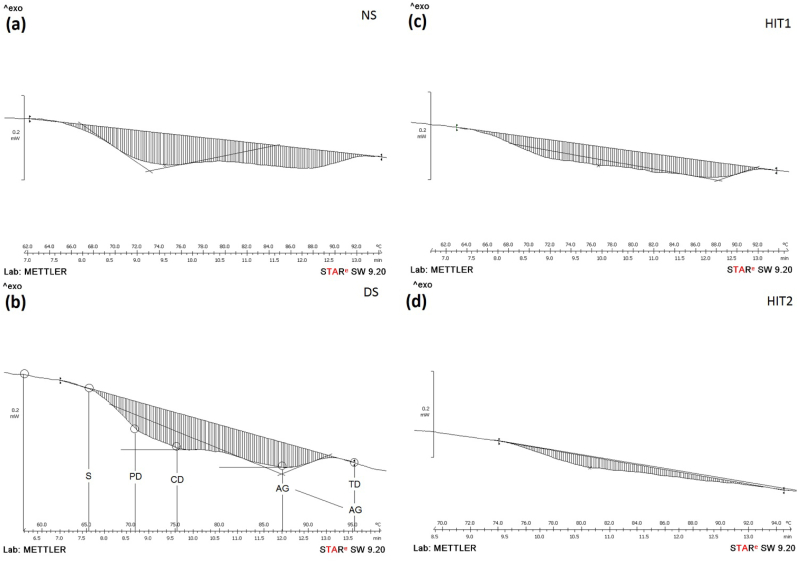
DSC thermograms of heat irradiation treatment (HIT1) (a) and heat ionization treatment (HIT2) (b) of winged bean proteins powders. DSC thermograms of winged bean protein-powders as sample controls using DNS (c) and of DS (d).
Table 5.
Regression coefficients, R2, p-value of lack of fit, the significance probability (p-value, t-ratio) of regression coefficients in final reduced second-order polynomial models of the final reduced models.
| Responses |
ΔH (%, Y1) |
ΔS (%, Y2) |
ΔCp (%, Y3) |
|||
|---|---|---|---|---|---|---|
| Coefficient | HIT1 | HIT2 | HIT1 | HIT2 | HIT1 | HIT2 |
| β0 | −1.43421 | 0.838606 | 0.023053 | 0.020568a | 0.096552 | 0.090432a |
| β1 | 0.00458 | −0.001248 | −0.000369 | −0.000202 | −0.000628 | 0.000334 |
| β2 | 0.11891 | 0.237095a | 0.000893 | 0.001140 | −0.001313 | – |
| β3 | 0.34548 | −0.100428 | – | −0.002012 | – | −0.020451a |
| 0.00032 | −0.000227a | 0.000007 | −0.000002 | 0.000014 | −0.000022a | |
| 0.00119 | −0.016667a | 0.000016 | −0.000057 | 0.000077a | – | |
| 0.00668 | −0.022102a | – | −0.000203a | – | −0.000365 | |
| β12 | −0.00160a | −0.001892a | −0.000022a | −0.000013 | −0.000049 | – |
| β13 | – | 0.004971a | – | 0.000061a | – | 0.000377a |
| β23 | −0.01100a | – | – | – | – | – |
| R2 | 0.618 | 0.953 | 0.478 | 0.907 | 0.374 | 0.898 |
| p-value | 0.018 | 0.000 | 0.026 | 0.000 | 0.196 | 0.000 |
| Lack of fit (p-value) | 0.064 | 0.162 | 0.621 | 0.284 | 0.051 | 0.362 |
| Main effects (p-values) | ||||||
| X1 | 0.951 | 0.895 | 0.722 | 0.190 | 0.111 | 0.629 |
| X2 | 0.134 | 0.005a | 0.374 | 0.311 | 0.083 | – |
| X3 | 0.247 | 0.147 | – | 0.072 | – | 0.001a |
| Quadratic effects (p-values) | ||||||
| X11 | 0.423 | 0.003a | 0.226 | 0.136 | – | 0.000a |
| X22 | 0.059 | 0.019a | 0.057 | 0.561 | – | – |
| X33 | 0.769 | 0.000a | – | 0.010a | – | 0.279 |
| Interaction effects (p-values) | ||||||
| X1X2 | 0.027a | 0.040a | 0.026a | 0.329 | 0.039a | – |
| X1X3 | – | 0.000a | – | 0.000a | – | 0.000a |
| X2X3 | 0.041a | – | – | – | – | – |
| Main effects (t-ratios) | ||||||
| X1 | −0.063 | −0.136 | −0.363 | −1.396 | 1.303 | 0.494 |
| X2 | 1.618 | 3.469 | 1.063 | 1.063 | 1.110 | – |
| X3 | 1.224 | −1.561 | – | −1.994 | – | −4.110 |
| Quadratic effects (t-ratios) | ||||||
| X11 | 0.831 | −3.743 | 1.266 | −1.609 | – | −4.780 |
| X22 | 2.102 | −2.753 | 2.075 | −0.599 | – | – |
| X33 | 0.302 | −5.256 | – | −3.083 | – | −1.125 |
| Interaction effects (t-ratios) | ||||||
| X1X2 | −2.558 | −2.328 | −2.487 | −1.020 | −1.223 | – |
| X1X3 | – | 7.340 | – | 5.767 | – | 7.180 |
| X2X3 | −2.313 | – | – | – | – | – |
| OP (X1, X2, X3) | ||||||
| OPHIT1 (68.5, 10, 2.56) | 1.1060 | – | 0.0154 | – | 0.0683 | – |
| OPHIT2 (40, 1.56, 2,5) | – | 0.7252 | – | 0.0100 | – | 0.0449 |
Abbreviation:β0 is a constant, βi, βii and βij are the linear, quadratic and interaction coefficients of the quadratic polynomial equation, respectively. i: Heating temperature generated via heat irradiation or heat ionization; ii: Time conducted for both treatments; ij: Mass of starting material for heat irradiation treatment or pH of buffer system for heat ionization.
Significant at p < 0.05.
The use of DSC is important in differentiating between non-mutant and mutant proteins, particularly at the beginning and at the end of the heating profiles (Fig. 5). In some literature review on cold adaptation of Atlantic salmon tropomyosin used differential scanning calorimetry DSC to monitor the thermal unfolding of mutant and non-mutant recombinant tropomyosin by direct heating. The observed melting temperature of the mutant ∼44 °C and non-mutant ∼40.5 °C protein differ by approximately 4 °C (Ige, 2012). This short range of temperature it can be expressed as well as into time per second. Same thing in our case we use time rather than temperature in expressing the index of stability. The index of stability is considered as new insights in the measurement of denaturation parameters where always from DSC we can observe or depict novel ideas like this one index of stability. The stability of proteins for a period of time expressed in seconds before undergoing denaturation gives an indication on the stability of the structure and intact form of the disulfide chemical bonds. This is new idea in DSC analysis that can be used in explaining the denaturation stages of protein. The thermal resistance of the protein powders before undergoing denaturation/unfolding using DSC circumstances provided strong evidences on the stability of proteins powders. This observation was very clear from the thermograms of DSC where HIT1 revealed an index of stability (S) of ∼30 s and of ∼54 s for HIT2. The 24 s was assigned as differences in the stability found in between HIT1 and HIT2 (Fig. 5 (c) and (d)). Thus, HIT2 was more stable than HIT1 based on denaturation/unfolding statuses and little bite speeded for HIT1, evidencing the role of thermal conductivity coefficient and to the penetrative power of the heat in between solid state and humid state of protein powders. These changes were attributed to the effect of high temperature short time coupled by pressure. These behaviours were associated as well as with the endothermic reaction that was mainly speeded protein agglomeration ability, gelatinization capacity and glassy state transitions. Partial denaturation PD was associated to the disordering effects in protein units and mainly reflected by the refolding of protein structure from quaternary to tertiary structure. For instance, complete denaturation CD indicated the disordering in the status of the polypeptides chains from tertiary and secondary forms to primary chain that were mainly accomplished by the breakdown of covalence bonds of disulfide chemical bridges and liberation of peptide chains. These effects gave initiation of aggregation AG domains where the formations of aggregates starts under reversible transition of chemical bonds. This stage was mainly ended by total denaturation TD domain where the protein units were totally segregated and denatured. Both of these mechanisms AG and TD could be displayed using DSC either via an extra shifting at the end of the profile or via an extra shifting at maximum temperatures. It was accepted that the aggregation mechanisms is one of the rearrangement aspects in polypeptide chain via reversible transition and usually takes an interval range from maximum temperature of denaturation (start of reversible transition) till end of the denaturation profile. The extra-shifting or exothermic shift at the end of the profile might depend on the type of protein and nature of experiment and its conditions. Thus, the fast denaturation levels was accompanied by extent level of protein aggregation e.g. by fast reconformation in polypeptide chains, whereas the slow denaturation level was attributed to the least effect of aggregation maintained by reversible chemical lesions. Fitzsimons, Mulvihill and Morris (2007) stated that the aggregation mechanism occurred slowly compared to the denaturation mechanism that was highly depending on the concentration of protein and ions strength used. Under these conditions both states can be visualised very well on the DSC profile.
Heat capacity ΔCp based model equation (3) was the maximum shifting in the thermograms that was mainly accompanied by reversible transition and aggregation of proteins and polypeptides. ΔCp was encrypted as well as in determining the heat flow velocity and solute transfers until a thermodynamic equilibrium status was maintained in between internal and external molecular forces. At this state the changes in the volume of protein polymeric chain was a strong evidence on the transition state of the energy and therefore on the ΔCp of denaturation/unfolding. The model parameters used in expressing the variation in the ΔCp are displayed in Table 5. As results, R2, significance of p-value, and insignificance of p-value of lack of fit for HIT1 were 0.374 (37.4%), 0.196, and 0.051, respectively. Whilst, the HIT2 demonstrated the following results of R2 of 0.898 (89.9%), p-value of 0.000, and p-value of lack of fit of 0.362. The obtained results were attributed to the differences in the activation energy of each method treatments and their capability in changing significantly protein structure motifs from complex units to simple ones. Thus, increasing the free rotational and movement of freely molecules within solutes changed the transitions states of molecules by rendering protein and associated water molecules in unfolded states (Cooper, Nutley and Walood, 2000; Bruylants, Wouters and Michaux, 2005; Wen et al., 2012). DSC thermograms of the control samples DS and NDS are shown in Fig. 5 (a) and (b). The results indicated that NDS was stable for a period time of 40.5 s, whereas the stability of DS was found to be 48 s. Almost both control samples showed a similar stability index with little difference of ∼7 s (Fig. 5). These results were attributed to the distribution of the amino acids capable for phosphorylation and able in forming disulfide chemical bridges. Another explanation was attributed to the thermal conductivity that was enhanced by tighted lipid to proteins unites where these fat globules were capable in rendering proteins in agglomerating states by speeding up the heat transfer earlier as compared to DS control. Gibbs free energy (ΔG) was used in determining protein denaturation (Edsall and Gutfreund, 1983; Breslauer, Freier and Straume, 1992; Krug, Hunter and Grieger, 1976; Gill, Moghadam and Ranjbar, 2010; Owusu-Apenten, 2005; Ford and Willson, 1999). The ΔG could be determined at the optimum point conditions of method treatments for HIT1 and HIT2.
3.7. Regression model- parameters of ΔH, ΔS, and ΔCp based significant interaction effects
Table 5 shows the significance in between factor interactions determined for protein denaturation/unfolding mechanism. HIT1 significantly changed the ΔH at the interaction effect of temperature and time and in between time and mass. The resulting p-value and t-ratio were 0.027 and −2.558 for temperature × time, and were 0.041 and −2.313 for time × mass, respectively. The denaturation was significant in terms of the ΔH to HIT2 by temperature × time and by temperature × pH. Their corresponding values of p-value and t-ratio for those interaction effects were 0.040 and −2.328 and were 0.00 and 7.340, respectively. The interaction effect for ΔS was found to be significant for HIT1at temperature × time with a p-value and a t-ratio of 0.026 and −2.487, respectively. HIT2 was found to be significant at interaction effects of temperature × pH with p-value and t-ratio of 0.00 and 5.767, respectively. The interaction effect of HIT1for ΔCp was found to be significant for temperature × time, resulting in p-value and t-ratio of 0.039 and −1.223, respectively. HIT2 revealed significant interaction effects with p-value of 0.00 and t-ratio of 7.180 for temperature × pH. The quadratic effect of ΔH was found to be significant for both method treatments. At molecular levels several conformational changes would take places such as the compaction and association of the resulting macromolecules and also to the aggregation in between polypeptide chains under specific and non-specific interaction forces or due to thermal unfolding of the remaining protease inhibitors that were mainly accomplished by formation of tetrameric aggregate forms that comprise disulfide interchanges (Pouvreau, Gruppen, van Koningsveld, van den Broek and Voragen, 2005). These changes were the results of heat exchange properties of the endo/exo material statuses and distribution of the components energy and ions within it (Galeb et al., 2012; Aguilera, 2000; Baer, Cassidy and Hiltner, 1992; Baianu, 1992; Privalov, Khechinashvili and Atanasov, 1971).
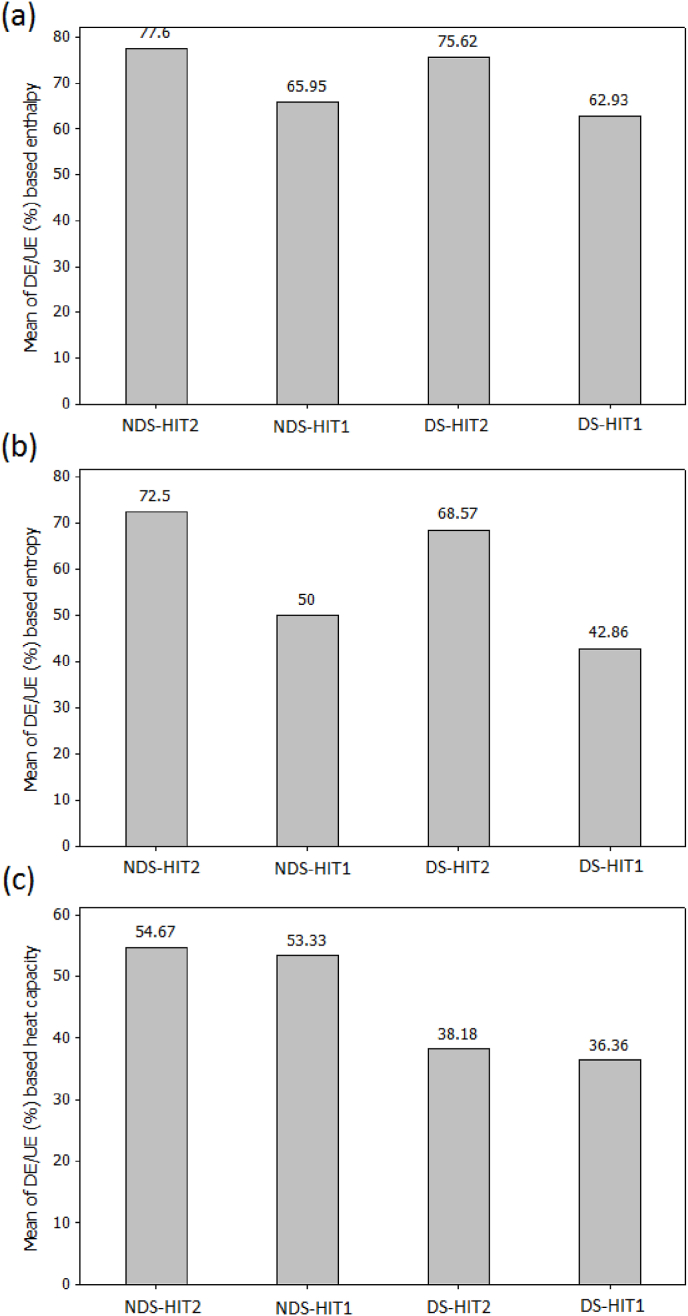
Table 5 shows the results of response optimizer regenerated at the optimum point of the treatment conditions of HIT1 and HIT2. The response variables were stratified according to factor ranges in the Minitab by selecting maximum, minimum and target point as levels of response variables. In the current study, the maximum level of the thermodynamic parameters of HIT1 and HIT2 of ΔH, ΔS, and ΔCp was selected in order to obtain a desirable point leading to a better contact surface exposure and therefore to the maximal denaturation/unfolding DE/UF% properties. Table 5, at significant interactions factors of 40 °C, 1.5663 h and 2.5 pH of HIT2 the generated optimum point using response optimizer for ΔH, ΔS, and ΔCp were 0.7252 J/g, 0.0100 mJ/k, and 0.0049 J/g × °C, respectively. The obtained optimum point for HIT1 conditions of temperature of 68.4486 °C, time of 10 min, and 2.5594 g as material mass were 1.1060 J/g, 0.0154 mJ/k, and 0.0683 J/g × °C for ΔH, ΔS, and ΔCp, respectively. The results of the DE/UF% of HIT2 determined in terms of ΔH and by using NDS and DS as controls yielded 77.6% and 75.62%, respectively, whereas for HIT1 and by using NDS and DS as controls the results were 65.95% and 62.93%, respectively. The obtained results determined in terms of ΔS for HIT2 and by using NDS and DS were 72.5% and 68.57%, respectively, whereas for HIT1 the results were 50% and 42.86%, respectively. In terms of ΔCp the DE/UE (%) using NDS and DS as controls were 54.67%, and 38.18%, respectively for HIT2, whereas for HIT1 the results were 53.33% and 36.36%, respectively (Fig. 6 (a), (b) and (c)). It was remarked that HIT2 efficiently denatured/unfolded the protein as compared to the impact of HIT1. The results of DE/UE (%) based ΔH, ΔS and ΔCp reflected the suitability and applicability of RSM model equations to structure reconformation and stability where the precision of the model equations was found to be in the following order ΔH> ΔS> ΔCp as shown in Fig. 6.
Fig. 6.
Changes in the denaturation/unfolding efficiency expressed in terms of enthalpy (a), entropy (b) and heat capacity (c).
4. Conclusion
The structural motif of winged bean seed proteins were successfully modified physically, chemically and enzymatically by respective heat ionization and heat irradiation treatments HIT1 and HIT2. Then the samples were followed by hydrolysis process that was enhanced successfully after denaturing/unfolding the protease inhibitors of BBI, KTI, lectin and by degrading other polyphenol compounds. The disordering in the molecular statuses led to significant reconformation in the e.g. molecular assembly. The exerting hydrolysis induced distinct transitions in the structures accomplished by the variation in the deviation angle (DA°) and folded/unfolded volumes. The applied method treatments induced reconformation and led to co-polymerization and aggregation of protein units by assigning variable range of denaturation/unfolding levels. This study has shown as well as that HIT2 had more impact on the degradation/inactivation of KTI, BBI and lectin. The pH of 2.5 of glycine-HCl buffer during an extended time of hydrolysis of 1.5 h (90 min) and an adequate heating temperature of 40 °C mitigated those antinutrients efficiently and therefore, induced the acceleration of enzymatic hydrolysis. The mechanism of hydrolysis was improved due to significant changes in the chemical features associated to the destruction of macromolecules backbone and their polymorphisms, breakdown of disulfide and hydrogen bonds, and degradation of PA and TA. The results of protein digestibility indicated significant changes in the three model types of proteins used. As results the deactivations level of KTI in the following trypsin-digests (TDJ) taking NS as control were ordered as follow: HIT2-TDJ (47.39%) > HIT1-TDJ (46.21%). In contrast, their deactivation levels for BBI in the following trypsin-α-chymotrypsin digests (TDJ-α-CHDJ) were ordered as follow: HIT2-(TDJ-α-CHDJ) 57.87% > HIT1-(TDJ-α-CHDJ) 30.23%.
CRediT authorship contribution statement
Sami Saadi: Writing – original draft, Writing – review & editing. Nazamid Saari: Supervision, editing and Validation. Hasanah Mohd Ghazali: Co-supervision, editing and Validation. Mohammed Sabo Abdulkarim: Co-supervision, editing and Validation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Authors acknowledge the Malaysian Ministry of Science, Technology and Innovation for the grant awarded to Professor Dr. Nazamid Saari under project number 10-05-ABI-PB038.
Contributor Information
Sami Saadi, Email: sami.saadi84@gmail.com, saadi.sami@ymail.com.
Nazamid Saari, Email: nazamid@upm.edu.my.
References
- Aguilera J.M. In: Food Preservation Technology Series: Trends in Food Engineering. Lozano J.E., Añón C., Parada-Arias E., Barbosa-Cánovas G.V., editors. Technomic Publishing Company, Inc; USA: 2000. Structure-property relationships in foods; p. 10. [Google Scholar]
- Alonso R., Orúe E., Marzo F. Effects of extrusion and conventional processing methods on protein and antinutritional factor contents in pea seeds. Food Chem. 1998;63:505–512. [Google Scholar]
- Anta L., Marina M.L., García M.C. Simultaneous and rapid determination of the anticarcinogenic proteins Bowman-Birk inhibitor and lectin in soybean crops by perfusion RP-HPLC. J. Chromatogr., A. 2010;1217:7138–7143. doi: 10.1016/j.chroma.2010.09.026. [DOI] [PubMed] [Google Scholar]
- Arinathan V., Mohan V.R., De Britto A.J. Chemical composition of certain tribal pulses in South India. Int. J. Food Sci. Nutr. 2003;54:209–217. doi: 10.1080/09637480120092026. [DOI] [PubMed] [Google Scholar]
- Arun A.B., Sridhar K.R., Raviraja N.S., Schmidt E., Jung K. Nutritional and antinutritional components of seeds of Canavalia spp. from the west coast sand dunes of India. Plant Foods Hum. Nutr. 2003;58:1–13. [Google Scholar]
- Baer E., Cassidy J.J., Hiltner A. In: ACS Symposium Series 489. Glasser W., Hatakeyama H., editors. American Chemical Society; Washington, DC: 1992. Hierarchical structure of collagen composite systems. In: viscoelasticity of biomaterials; pp. 2–23. [Google Scholar]
- Baianu I.C. Physical Chemistry of Food Processes. Fundamental Aspects. Van Nostrand Reinhold; New York: 1992. Structural techniques; p. 200. [Google Scholar]
- Barman A., Marak C.M., Barman R.M., Sangma C.S. In: Legume Seed Nutraceutical Research. Jimenez-Lopez J.C., Clemente A., editors. IntechOpen; London, UK: 2018. Nutraceutical properties of legume seeds and their impact on human health, legume seed nutraceutical research. [Google Scholar]
- Boye J., Zare F., Pletch A. Pulse proteins: processing, characterization, functional properties and applications in food and feed. Food Res. Int. 2010;43:414–431. [Google Scholar]
- Brenes A., Jansman A.J.M., Marquardt R.R. In: Recent Advances of Research in Antinutritional Factors in Legume Seeds and Oilseeds. Muzquiz M., Hill G.D., Cuadrado C., Pedrosa M.M., Burbano C., editors. Wageningen Academic Publishers; Wageningen: 2004. Recent progress on research on the effects of antinutritional factors in legume and oil seeds in monogastric animals; pp. 195–217. [Google Scholar]
- Breslauer K.J., Freier E., Straume M. Calorimetry: a tool for DNA and ligand-DNA studies. Methods Enzymol. 1992;211:533–567. doi: 10.1016/0076-6879(92)11030-m. [DOI] [PubMed] [Google Scholar]
- Bruylants G., Wouters J., Michaux C. Differential scanning calorimetry in life science: thermodynamics, stability, molecular recognition and application in drug design. Curr. Medicine Chem. 2005;12:2011–2020. doi: 10.2174/0929867054546564. [DOI] [PubMed] [Google Scholar]
- Carbonaro M. In: Pulse Foods. Tiwari B.K., Gowen A., McKenna B., editors. Academic Press; San Diego, CA, USA: 2011. 14-Role of pulses in nutraceuticals; pp. 385–418. [Google Scholar]
- Cerny K., Korydylas M., Pospisil F., Svabensky O., Zajir B. Nutritive value of the winged bean (Psophocarpus palustris Desv.) Br. J. Nutr. 1971;26:293. doi: 10.1079/bjn19710035. [DOI] [PubMed] [Google Scholar]
- Champ M.M.J. Non-nutrient bioactive substances of pulses. Br. J. Nutr. 2002;88:S307–S319. doi: 10.1079/BJN2002721. [DOI] [PubMed] [Google Scholar]
- Church F.C., Swaisgood H.E., Porter D.H., Catignani G.L. Spectrophotometric assay using O-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J. Dairy Sci. 1983;66:1219–1227. [Google Scholar]
- Clawson G.A. Protease inhibitors and carcinogenesis: a review. Cancer Invest. 1996;14:597–608. doi: 10.3109/07357909609076904. [DOI] [PubMed] [Google Scholar]
- Clemente A., Gee J.M., Johnson I.T., MacKenzie D.A., Domoney C. Pea (Pisum sativum L.) protease inhibitors from the Bowman Birk class influence the growth of human colorectal adenocarcinoma HT29 cells in vitro. J. Agric. Food Chem. 2005;53:8979–8986. doi: 10.1021/jf051528w. [DOI] [PubMed] [Google Scholar]
- Cooper A., Nutley M.A., Walood A. In: Protein-Ligand Interactions: Hydrodynamics and Calorimetry. Harding S.E., Chowdhry B.Z., editors. Oxford University Press; Oxford, UK: 2000. Differential scanning microcalorimetry; pp. 287–318. [Google Scholar]
- D'Mello J.P.F. In: Tropical Legumes in Animal Nutrition. D'Mello J.P.F., Devendra C., editors. CAB International; Wallingford: 1995. Anti-nutritional substances in legume seeds; pp. 135–172. [Google Scholar]
- Dahimi O., Rahim A.A., Abdulkarim S.M., Hassan M.S., Zam Hashari B.T.S., Mashitoh A.S., Saadi S. Multivariate statistical analysis treatment of DSC thermal properties for animal fat adulteration. Food Chem. 2014;158(1):132–138. doi: 10.1016/j.foodchem.2014.02.087. [DOI] [PubMed] [Google Scholar]
- Deshpande S.S., Damodaran S. Structure-digestibility relationship of legume 7S proteins. J. Food Sci. 1989;54(1):108–113. [Google Scholar]
- Domoney C. In: Seed Protein. Shewry P.R., Casey R., editors. Amsterdam Kluwer Academic Publishers; 1999. Inhibitor of legume seeds; pp. 635–655. [Google Scholar]
- Edsall T.J., Gutfreund H. In: Biothermodynamics: the Study of Biochemical Processes at Equilibrium. Edsall T.J., Gutfreund H., editors. John Wiley & Sons; New York, NY, USA: 1983. Calorimetry, heat capacity, and phase transitions; pp. 210–227. [Google Scholar]
- Feil B. Phytic acid. J. N. Seeds. 2001;3:1–35. [Google Scholar]
- Fitzsimons S.M., Mulvihill D.M., Morris E.R. Denaturation and aggregation processes in thermal gelation of whey proteins resolved by differential scanning calorimetry. Food Hydrocolloids. 2007;21:638–644. [Google Scholar]
- Ford J.L., Willson B.R. In: Kemp R., editor. vol. 4. Elsevier; The Netherlands: 1999. Thermal analysis and calorimetry of pharmaceuticals; pp. 923–1016. (Handbook of Thermal Analysis and Calorimetry). [Google Scholar]
- Fredrikson M., Biot P., Alminger M., Carlsson N., Sandberg A. Production process for high-quality pea-protein isolate with low content of oligosaccharides and phytate. J. Agric. Food Chem. 2001;49:1208–1212. doi: 10.1021/jf000708x. [DOI] [PubMed] [Google Scholar]
- Galeb H.A., Salimon J., Eid E.E.M., Nacer N.E., Saari N., Saadi S. The impact of single and double hydrogen bonds on crystallization and melting regimes of Ajwa and Barni lipids. Food Res. Int. 2012;48:657–666. [Google Scholar]
- Garcia V.V., Palmer J.K. Proximate analysis of five varieties of winged beans Psophocarpus tetragonolobus (L.) DC. J. Food Technol. 1980;15:469–476. [Google Scholar]
- Garg S.K., Makkar H.P.S., Nagal K.B., Sharma S.K., Wadhwa D.R., Singh B. Toxicological investigations into oak (Quercus incana) leaf poisoning in cattle. Vet. Hum. Toxicol. 1992;34:161–164. [PubMed] [Google Scholar]
- Ghanbari R., Ebrahimpour A., Abdul-Hamid A., Ismail A., Saari N. Actinopyga lecanora hydrolysates as natural antibacterial agents. Int. J. Mol. Sci. 2012;13:16796–16811. doi: 10.3390/ijms131216796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giami S.Y., Bekebain D.A., Emelike N.J.T. Proximate composition and functional properties of winged bean (Psophocarpus tetragonolobus) Niger. J. Nutri. Sci. 1992;13:35–39. [Google Scholar]
- Gill P., Moghadam T.T., Ranjbar B. Differential scanning calorimetry techniques: applications in biology and nanoscience. J. Biomol. Tech. 2010;21:167–193. [PMC free article] [PubMed] [Google Scholar]
- Henley E.C., Kuster J.M. Protein quality evaluation by protein digestibility-corrected amino acid scoring. Food Technol. 1994;4:74–77. [Google Scholar]
- Hurrell R.F., Juillerat M.A., Reddy M.B., Lynch S.R., Dassenko S.A., Cook J.D. Soy protein, phytate, and iron absorption in humans. Am. J. Clin. Nutr. 1992;56:573–578. doi: 10.1093/ajcn/56.3.573. [DOI] [PubMed] [Google Scholar]
- Hussain I., Uddin M.B., Aziz M.G. Optimization of antinutritional factors from germinated wheat and mungbean by response surface methodology. Int. Food Res. J. 2011;18(3):957–963. [Google Scholar]
- Ige T.O. Department of Biochemistry, Memorial University of Newfoundland St John’s N.L Canada; 2012. Cold Adaptation of Atlantic Salmon Tropomyosin: Role of Residue 77. Thesis of Master of Science; pp. 1–119. [Google Scholar]
- Jamroz D., Kubizna J. Harmful substances in legume seeds - their negative and beneficial properties. Pol. J. Vet. Sci. 2008;11:389–404. [PubMed] [Google Scholar]
- Kayodé A.A.P., Nout M.J.R., Bakker E.J., Van Boekel M.A.J.S. Evaluation of the simultaneous effects of processing parameters on the iron and zinc solubility of infant sorghum porridge by response surface methodology. J. Agric. Food Chem. 2006;54:4253–4259. doi: 10.1021/jf0530493. [DOI] [PubMed] [Google Scholar]
- Krug R.R., Hunter W.G., Grieger R.A. Enthalpy-entropy compensation. 1. Some fundamental statistical problems associated with the analysis of van't Hoff and Arrhenius data. J. Phys. Chem. 1976;80:2335–2341. [Google Scholar]
- Kumar V., Sinha A.K., Makkar H.P.S., Becker K. Dietary roles of phytate and phytase in human nutrition: a review. Food Chem. 2010;120:945–959. [Google Scholar]
- Laskowsky M., Kato I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- Lee J., Ye L., Landen W.O., Eitenmiller R.R. Optimization of an extraction procedure for quantification of vitamin E in tomato and broccoli using response surface 13-methodology. J. Food Compos. Anal. 2000;13:45–57. [Google Scholar]
- Lee J.-E., Bae I.Y., Lee H.G., Yang C.-B. Tyr-Pro-Lys, an angiotensin I-converting enzyme inhibitory peptide derived from broccoli (Brassica oleracea Italica) Food Chem. 2006;99:143–148. [Google Scholar]
- Liener I.E. In: Recent Advances of Research in Antinutritional Factors in Legume Seeds. Huisman J., van der Poel A.F.B., Liener I.E., editors. Centre for Agricultural Publishing and Documentation (PUDOC); Wageningen: 1989. Antinutritional factors in legume seeds: state of the art; pp. 6–13. [Google Scholar]
- Liu B.L., Rafiq A., Tzeng Y.M., Rob A. The induction and characterization of phytase and beyond. Enzym. Microb. Technol. 1998;22:215–424. [Google Scholar]
- Maga J.A. Phytate: its chemistry, occurrence, food interactions, nutritional significance, and methods of analysis. J. Agric. Food Chem. 1982;30:1–9. [Google Scholar]
- Maphosa Y., Jideani V.A. In: Functional Food—Improve Health through Adequate Food. Hueda M.C., editor. IntechOpen; London, UK: 2017. The role of legumes in human nutrition. [Google Scholar]
- Mirhosseini H., Tan C.P., Taherian A.R., Boo H.C. Modeling the physicochemical properties of orange beverage emulsion as function of main emulsion components using response surface methodology. Carbohydr. Polym. 2009;75:512–520. [Google Scholar]
- Montgomery D.C. Wiley; New York: 2001. Design and Analysis of Experiments. [Google Scholar]
- Morrison S.C., Savage G.P., Morton J.D., Russell A.C. Identification and stability of trypsin inhibitor isoforms in pea (Pisum sativum L.) cultivars grown in New Zealand. Food Chem. 2007;100:1–7. [Google Scholar]
- Mosolov V.V., Valueva T.A. Proteinase inhibitors and their function in plants: a review. Appl. Biochem. Microbiol. 2005;41:227–246. [PubMed] [Google Scholar]
- Muzquiz M., Varela A., Burbano C., Cuadrado C., Guillamón E., Pedrosa M.M. Bioactive compounds in legumes: pronutritive and antinutritive actions. Implications for nutrition and health. Phyt. Rev. 2012;11:227–244. [Google Scholar]
- Myers R., Montgomery D.C. Wiley; New York: 2002. Response Surface Methodology: Process and Product Optimization Using Designed Experiments. [Google Scholar]
- Owusu-Apenten R.K. . CRS Press; Boca Raton London New York Washington, D.C: 2005. Chemistry and Food Chemistry: an Overview in: Introduction to Food Chemistry; pp. 4–7. [Google Scholar]
- Oomah B.D., Cardadro-Martínez A., Loarca-Piña G. Phenolics and antioxidative activities in common beans (Phaseolus vulgaris L.) J. Sci. Food Agric. 2005;85:935–942. [Google Scholar]
- Pedrosa M.M., Guillamón E., Arribas C. Autoclaved and extruded legumes as a source of bioactive phytochemicals: a review. Foods. 2021;10:379. doi: 10.3390/foods10020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouvreau L., Gruppen H., van Koningsveld G., van den Broek L.A.M., Voragen A.G.J. Conformational stability of the potato serine protease inhibitor group. J. Agric. Food Chem. 2005;53(8):3191–3196. doi: 10.1021/jf048353v. [DOI] [PubMed] [Google Scholar]
- Prakash D., Misra P.N., Misra P.S. Amino acid profile of winged bean (Psophocarpus tetragonolobus) (L.): a rich source of vegetable protein. Plant Foods Hum. Nutr. 1987;37:261–264. doi: 10.1007/BF01091791. [DOI] [PubMed] [Google Scholar]
- Privalov P.L., Khechinashvili N.N., Atanasov B.P. Thermodynamic analysis of thermal transitions in globular proteins. I. Calorimetric study of chymotrypsinogen, ribonuclease, and myoglobin. Biopolymers. 1971;10:1865–1890. doi: 10.1002/bip.360101009. [DOI] [PubMed] [Google Scholar]
- Pusztai A., Bardocz S., Martín-Cabrejas M.A. In: Recent Advances of Research in Antinutritional Factors in Legume Seeds and Oilseeds. Muzquiz M., Hill G.D., Cuadrado C., Pedrosa M.M., Burbano C., editors. Wageningen Academic Publishers; Wageningen: 2004. The mode of action of ANFs on the gastrointestinal tract and its microflora; pp. 87–100. [Google Scholar]
- Qi R.-F., Song Z.-W., Chi C.-W. Structural features and molecular evolution of Bowman Birk protease inhibitors and their potential application. Acta Biochim. Biophys. 2005;37:283–292. doi: 10.1111/j.1745-7270.2005.00048.x. [DOI] [PubMed] [Google Scholar]
- Raboy V. Seed total phosphate and phytic acid. Biotechnol. Agric. For. 2009;63:41–53. [Google Scholar]
- Rokka T., Syväoja E.-L., Tuominen J., Korhonen H. Release of bioactive peptides by enzymatic proteolysis of Lactobacillus GG fermented UHT-milk. Milchwissenschaft. 1997;52:675–678. [Google Scholar]
- Roy F., Boye J.I., Simpson B.K. Bioactive proteins and peptides in pulse crops: pea, chickpea and lentil. Food Res. Int. 2010;43:432–442. [Google Scholar]
- Saadi S., Ariffin A.A., Ghazali H.M., Abdulkarim M.S., Boo H.C., Miskandar M.S. Crystallisation regime of w/o emulsion [e.g. multipurpose margarine] models during storage. Food Chem. 2012;133:1485–1493. [Google Scholar]
- Saadi S., Ariffin A.A., Ghazali H.M., Miskandar M.S., Abdulkarim S.M., Boo H.C. Effect of blending and emulsification on thermal behavior, solid fat content, and microstructure properties of palm oil- based margarine fats. J. Food Sci. 2011;76(1):C21–C30. doi: 10.1111/j.1750-3841.2010.01922.x. [DOI] [PubMed] [Google Scholar]
- Saadi S., Ariffin A.A., Ghazali H.M., Miskandar M.S., Boo H.C., Abdulkarim S.M. Application of differential scanning calorimetry (DSC), HPLC, and pNMR for interpretation primary crystallisation caused by combined low and high melting TAGs. Food Chem. 2012;132:603–612. doi: 10.1016/j.foodchem.2011.10.095. [DOI] [PubMed] [Google Scholar]
- Saadi S., Saari N., Abdulkarim S.M., Ghazali H.M., Anwar F. Smart electrical bi-layers lipopeptides: novel peptidic chains like zigzag map esterified with phospho-glyceride as mono-layer moieties capable in forming a meso-sphere- envelop with scaffold ability to cellular impurities. Control. Release. 2018;274:93–101. doi: 10.1016/j.jconrel.2017.10.011. [DOI] [PubMed] [Google Scholar]
- Saadi S., Saari N., Farooq A., Abdul Hamid A., Ghazali H.M. Recent advances in food biopeptides: production, biological functionalities and therapeutic applications. Biotechnol. Adv. 2015;33(1):80–116. doi: 10.1016/j.biotechadv.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Saiga A., Okumura T., Makihara T., Katsuda S.I., Morimatsu F., Nishimura T. Action mechanism of an angiotensin I-converting enzyme inhibitory peptide derived from chicken breast muscle. J. Agric. Food Chem. 2006;54:942–945. doi: 10.1021/jf0508201. [DOI] [PubMed] [Google Scholar]
- Saini H.S. Thermal stability of protease inhibitors in some cereals and legumes. Food Chem. 1989;32:59–67. [Google Scholar]
- Salami M., Yousefi R., Ehsani M.R., Dalgalarrondo M., Chobert J.M., Haertlé T., Razavi S.H., Saboury A.A., Niasari-Naslaji A., Moosavi-Movahedi A.A. Kinetic characterization of hydrolysis of camel and bovine milk proteins by pancreatic enzymes. Int. Dairy J. 2008;18:1097–1102. [Google Scholar]
- Sauveur D.S., Gauthier S.F., Boutin Y., Montoni A. Immunomodulating properties of a whey protein isolate, its enzymatic digest and peptide fractions. Int. Dairy J. 2008;18:260–270. [Google Scholar]
- Schlemmer U., Frølich W., Prieto R.M., Grases F. Phytate in foods and significance for humans: food sources, intake, processing, bioavailability, protective role and analysis. Mol. Nutr. Food Res. 2009;53:S330–S375. doi: 10.1002/mnfr.200900099. [DOI] [PubMed] [Google Scholar]
- Singh B., Singh J.P., Shevkani K., Singh N., Haur A. Bioactive constituents in pulses and their health benefits. J. Food Sci. Technol. 2017;54:858–870. doi: 10.1007/s13197-016-2391-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangendjaja B., Buckle K.A., Wootton M. Analysis of phytic acid by high-performance liquid chromatography. J. Chromatogr. 1980;197:274–277. [Google Scholar]
- Terrill T.H., Waghorn G.C., Woolley D.J., McNabb W.C., Barry T.N. Assay and digestion of 14C-labelled condensed tannins in the gastrointestinal tract of sheep. Br. J. Nutr. 1994;72:467–477. doi: 10.1079/bjn19940048. [DOI] [PubMed] [Google Scholar]
- USA . vol. 2011. U.S. Department of Health and Human Services; Washington, DC: 2005. (US Dietary Guidelines for Americans). [Google Scholar]
- Verzele M., Delahaye P. Analysis of tannic acids by high performance liquid chromatography. J. Chromatogr. 1983;268:469–476. [Google Scholar]
- Wang H.X., Ng T.B. An antifungal peptide from red lentil seeds. Peptides. 2007;28(3):547–552. doi: 10.1016/j.peptides.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Welch R. The impact of mineral nutrients in food crops on global human health. Plant Soil. 2002;247(1):83–90. [Google Scholar]
- Wen J., Arthur K., Chemmalil L., Muzammil S., Gabrielson J., Jiang Y. Applications of differential scanning calorimetry for thermal stability analysis of proteins: qualification of DSC. J. Pharm. Sci. 2012;101(3):955–996. doi: 10.1002/jps.22820. [DOI] [PubMed] [Google Scholar]
- Ye X.Y., Ng T.B. Isolation of a new cyclophilin-like protein from chickpeas with mitogenic, antifungal, and anti-HIV-1 reverse transcriptase activities. J. Life Sci. 2002;70(10):1129–1138. doi: 10.1016/s0024-3205(01)01473-4. [DOI] [PubMed] [Google Scholar]
- Yea C.S., Ebrahimpour A., Abdul-Hamid A., Bakar J., Muhammad K., Saari N. Winged bean [Psophocarpus tetragonolobus (L.) DC] seeds as an underutilized plant source of bifunctional proteolysate and biopeptides. Food Funct. 2014;5:1007–1016. doi: 10.1039/c3fo60667h. [DOI] [PubMed] [Google Scholar]
- Zarei M., Ebrahimpour A., Hamid A.A., Farooq A., Nazamid S. Production of defatted palm kernel cake protein hydrolysate as a valuable source of natural antioxidant. Int. J. Mol. Sci. 2012;13:8097–8111. doi: 10.3390/ijms13078097. [DOI] [PMC free article] [PubMed] [Google Scholar]