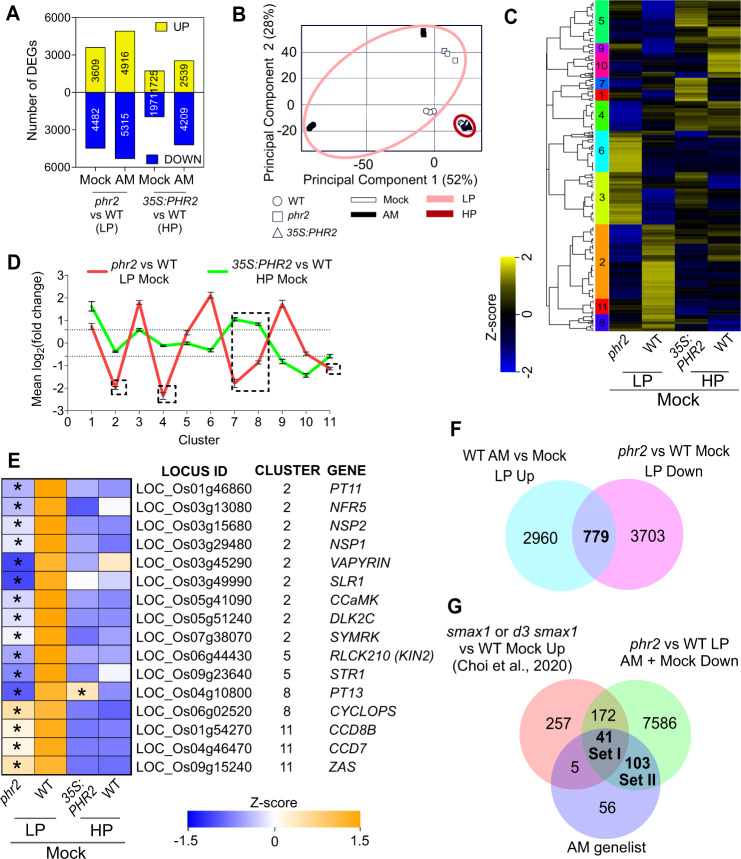
Fig. 2. The PHR2-dependent transcriptome of non-colonized roots contains genes required for AM development.
A Number of up- and downregulated differentially expressed genes (DEGs) in phr2 vs WT (LP) and 35S:PHR2 vs WT (HP) for mock or R. irregularis-inoculated plants. B PCA plot for the transcriptome of indicated samples. C Hierarchical clustering of combined DEGs from mock-inoculated roots using normalized counts. Colored bars on the left of the heatmap depict individual clusters (based on the dendrogram). Z-scores represent scaled normalized counts. D Mean log2FC of phr2 vs WT (LP) and 35S:PHR2 vs WT (HP) for clusters obtained in (C). Dotted lines indicate mean log2FC values of −0.585 and 0.585 (used as a cut-off for selecting DEGs). Dashed boxes highlight the mean log2FC of clusters with overall reduced transcript accumulation in phr2 vs WT. E Z-scores for a subset of genes from (C) previously reported being functionally required in AM. Asterisks indicate that the gene is a DEG in the phr2 vs WT (LP) or 35S:PHR2 vs WT (HP) comparisons. F Venn diagram showing overlap of DEGs with increased transcript accumulation in wild type ‘AM vs Mock’ and DEGs with reduced transcript accumulation in ‘phr2 vs WT’ in Mock at LP. G Venn diagram showing the intersection of DEGs with reduced transcript accumulation in the ‘phr2 vs WT’ comparison for mock and AMF-inoculated samples with AM genelist (Supplementary Data 4) and DEGs upregulated in either smax1 (vs WT) or d3 smax1 (vs WT) Mock24. Statistics: Mean ± SE are shown in (D) where N = Number of genes in each cluster as obtained in (C). Error bars represent the SE of the mean.