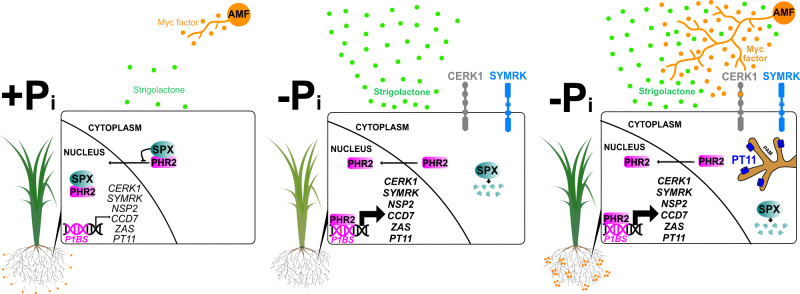
Fig. 6. Model depicting regulation of AM symbiosis by PHR2.
When plants obtain sufficient phosphate (left), SPX proteins prevent nuclear translocation of PHR242, as well as PHR2 binding to promoters of phosphate starvation -induced genes including AM relevant genes17,42. This causes low exudation of strigolactone and poor expression of genes required for perception of Myc-Factors and fungal entry, thereby preventing full symbiosis development. Upon phosphate starvation, SPX proteins, are degraded42. Consequently, PHR2 is active, can bind to P1BS elements in promoters, and transcriptionally activate genes important for AM, such as CCD7 involved in strigolactone biosynthesis for activation of the fungus in the rhizosphere prior to contact31–33, genes encoding receptors involved in the perception of fungal signals prior to root contact such as CERK1 and SYMRK39,40,65,66, the transcription factor NSP267, ZAS involved in apocarotenoid biosynthesis promoting root colonization68, and the AM-specific phosphate transporter gene PT1141 (localized to the peri-arbuscular membrane (PAM)) required for Pi uptake from the fungus. (For simplicity, we focus here on 6 genes with important and genetically determined roles in AM that have been recovered in both ChIP-Seq replicates Fig. 3C, and were confirmed by ChIP-qPCR, Supplementary Fig. 17). Consequently, at low phosphate, roots exude increased amounts of strigolactone and can perceive fungal signals (middle), the fungus is activated to colonize the roots and the symbiosis can function through nutrient transporters localizing to the peri-arbuscular membrane (right). Thus, symbiosis establishment appears to be enabled as a part of the PHR2-regulated phosphate starvation response.