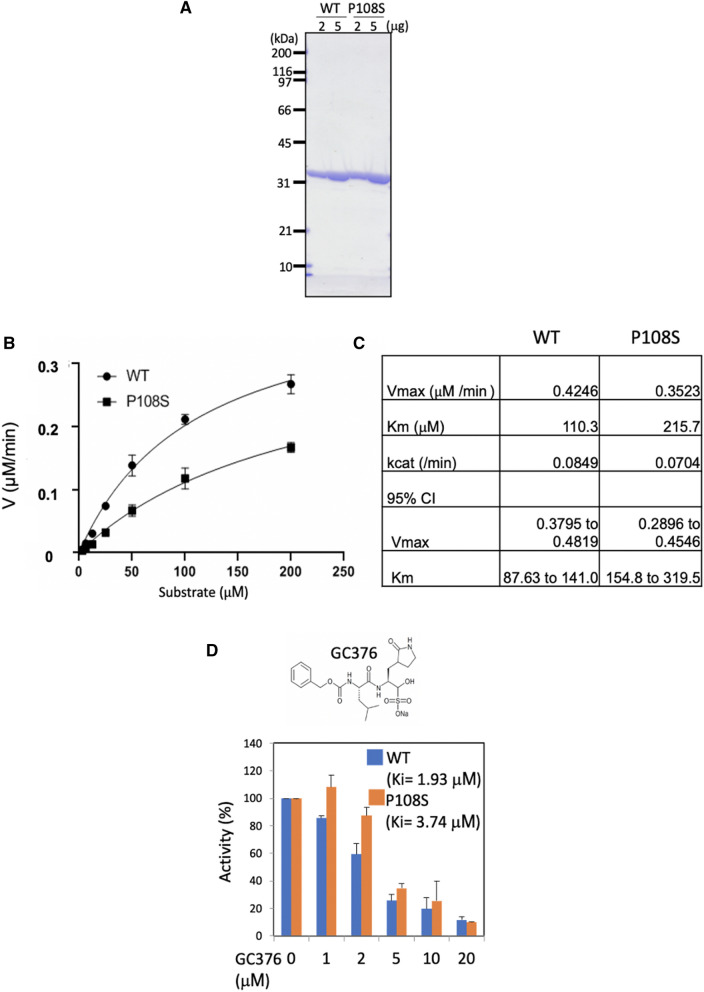
Figure 4.
SARS-CoV-2 3CLpro P108S is declined its enzymatic activity by structural alteration. (a) Recombinant WT or P108S of SARS-CoV2 3CLpro were analysed with SDS-PAGE visualizing using CBB staining. (b) The enzymatic activities of SARS-CoV2 3CLpro WT (circle) and P108S (square) were determined using a FRET-based substrate with the cleavage site of SARS CoV-2 3CLpro (Dabcyl-KTSAVLQ↓SGFRKME-Edans). Error bars show mean ± SD (n = 3). (c) The kinetic parameters of enzyme activity of 3CLpro WT and P108S were determined using GraphPad Prism 8 software by initial rate measurement of the substrate cleavage. The Kcat/Km value of the P108S mutant enzyme was 42% of that of the WT enzyme, showing 58% reduction. CI indicates 95% confidence interval. (d) Inhibitory activities of SARS-CoV2 3CLpro WT and P108S by GC376 were analyzed using a FRET-based cleavage assay. The graph shows the relative enzymatic activity. The inhibitory constant (Ki) was calculated using GraphPad Prism 8 software. Error bars show mean ± SD (n = 3). SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; 3CLpro, 3 chymotrypsin-like protease; WT, Wuhan-strain type; P108S, Pro108Ser-strain type; SDS-PAGE, Sodium dodecyl sulfate–Polyacrylamide gel electrophoresis; CBB, Coomassie Brilliant Blue; FRET, fluorescence resonance energy transfer.