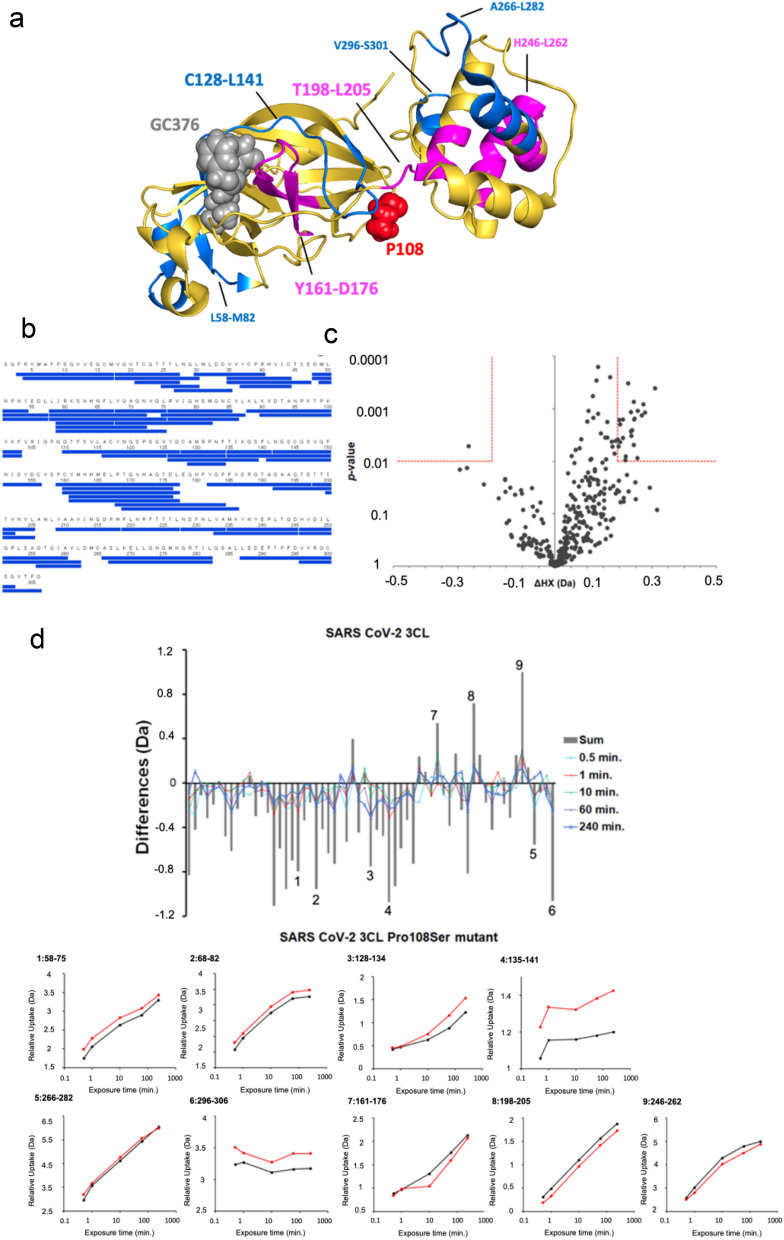
Figure 6.
HDX-MS results of SARS CoV-2 3CLpro WT and P108S. (a) Structurally influenced regions accompanied by a single mutation at 108th amino acid from proline to serine. HDX-MS showed more protected regions (magenta) and more exposed regions (cyan) in SARS-CoV-2 3CLpro Pro108Ser mutant compared to SARS-CoV-2 3CLpro. Mutation of proline to serine at 108th amino acid induces structural alternation at the regions from C128 to L141 and from Y161 to D176, where C128-L141 is sandwiched between P108 and Y161-D176 which is located at the substrate binding region. (b) The coverage map of identified peptides in SARS-CoV-2 3CLpro. (c) Volcano plots of observed delta HDX values and p values calculated from Welch’s t-test for SARS-CoV-2 3CL WT. Red lines showed the horizontal p value and the vertical delta HDX values for the significant criteria. (d) Differential plots of deuterium uptake degrees of peptides, showing time courses, along with their summational results (gray bar). Deuterium uptake curves for the peptides showing significant differences between WT (black) and P108S (red) proteins were presented. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; 3CLpro, 3 chymotrypsin-like protease; WT, Wuhan-strain type; P108S, Pro108Ser mutant. HDX-MS, Hydrogen/deuterium exchange mass spectrometry.