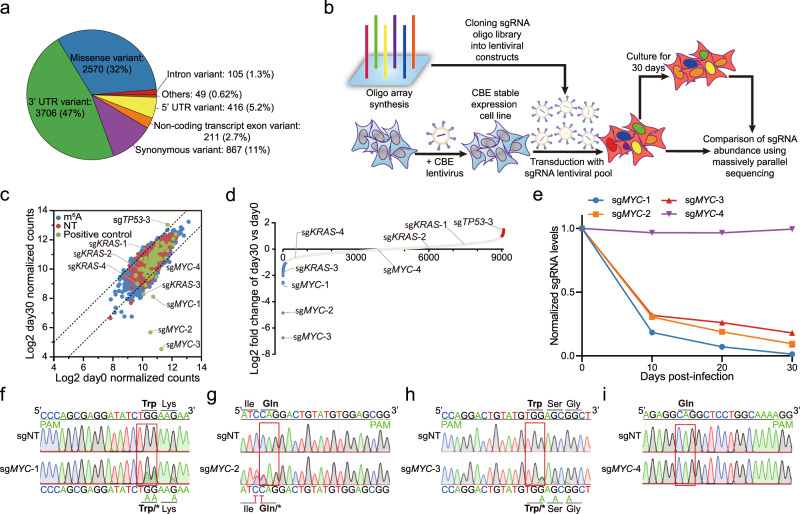
Fig. 2. FNLS-BE3 base editor-based screening in A549 cells.
a Variant effect prediction of FNLS-ABE7.10(AW) base editor-induced mutations for targetable loci. b Schematic illustration depicting FNLS-BE3 base editor-based screening in A549 cells. c Scatter plot comparing the normalized sgRNA counts of day 0 and day 30 during FNLS-BE3 base editor-based screening in A549 cells. The dotted lines indicate log2-fold change (LFC) of 1 and −1, respectively. d Dot plot showing the sorted LFCs of sgRNAs in FNLS-BE3 base editor-based screening in A549 cells. sgRNAs with LFC < −1 or LFC > 1 were colored in blue or red, respectively. e Relative abundances of MYC targeting sgRNAs predicted to induce BE3-mediated CRISPR-STOP during the expansion of A549 cells (n = 2 biologically independent samples). Data are presented as means. f–i Representative sequence chromatogram of FNLS-BE3-treated A549 cells at MYC-1 (f), MYC-2 (g), MYC-3 (h), and MYC-4 (i) loci. Source data are provided as a Source Data file.