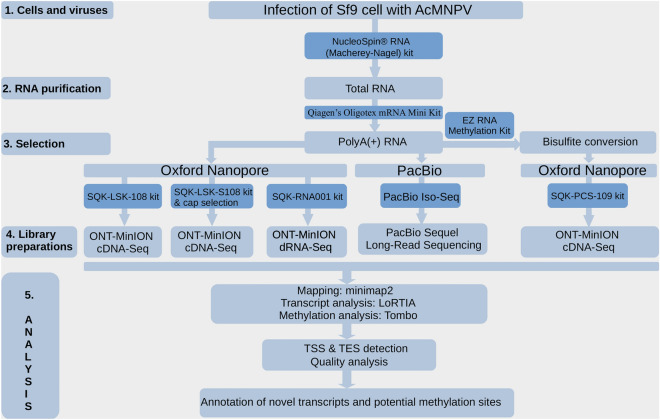
Figure 1.
Workflow of transcriptome and epitranscriptome analyses. The types of kits used for each workflow are shown in the dark blue boxes. Spodoptera frugiperda cells were infected with Autographa californica nuclear polyhedrosis virus (AcMNPV). RNA was isolated from the infected cells, and after poly (A) selection, the samples were sequenced using the Oxford Nanopore technologies (ONT) and the Pacific Biosciences (PacBio) Sequel platforms. In the case of ONT, we used PCR-amplified cDNA-seq (SQK-LSK-108, SQK-PCS-109 kit) and dRNA-seq (SQK-RNA001 kit). ONT dRNA-seq was also applied to determine possible methylation sites using the Tombo software. To confirm the methylation sites, bisulfite conversion was also performed using ONT sequencing. The base-called sequences were aligned with the reference genome using the minimap2 long-read mapping software, followed by the determination of TSS and TES positions of each transcript using the LoRTIA program. The illustration was created with Microsoft PowerPoint 2021 software47.