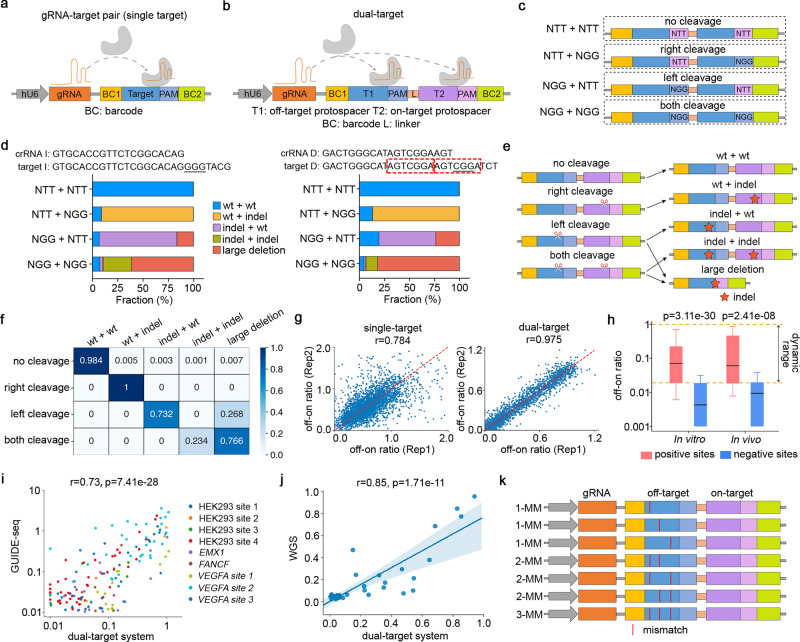
Fig. 1. Assessment of off-on ratios using a synthetic dual-target system.
a, b A schematic representation of paired gRNA-target design using a single-target or b dual-target sequence. c The design of control dual-target sequences corresponding to 4 cleavage types, where the left and right protospacers are identical and the cleavage events are turned on or off by “NGG” or “NTT” PAM sequences, respectively. d Bar charts showing the fractions of five types of editing outcomes in relation to the 4 cleavage types. Left: a gRNA associated with dominating non-homologous end joining (NHEJ); right: a gRNA associated with dominating microhomology-mediated end joining (MMEJ), where the microhomology sequences are highlighted in red rectangles. e A demonstration of the cleavage-editing model of the dual-target system. f A heatmap showing the cleavage-editing transition matrix computed from mutational profiles of 276 control dual-target sequences. g Scatter plots showing the correlations of off-on ratios between biological replicates using single- or dual-target systems. h A box plot showing the distributions of the off-on ratios estimated by the dual-target system, corresponding to the 32 filtered benchmark gRNAs and 480 target sequences. Positive: reported off-target sequences; Negative: unreported genomic sequences with <5 mismatches relative to crRNA. The data represent n = 204 positive, 213 negative (in vitro), and n = 66 positive, 52 negative (in vivo) gRNA-target pairs. The box plot displays a median line, interquartile range boxes and min to max whiskers. The p-values were calculated using two-tailed Manny–Whitney U test. i, j Scatter plots showing the correlations between the off-on ratios estimated from the dual-target system and i GUIDE-seq or j WGS, at the reported genomic off-target sequences. The p-values were calculated using Pearson correlation test. The shadow represents the 95% confidence interval. k A demonstration of the off-target sequence design in the high-throughput experiments using the dual-target synthetic system, where the mismatches in the 2-MM and 3-MM sequences are the combinations of the mismatches in the 1-MM sequences. Source data for Fig. 1d, g–j are provided in the Source Data file.