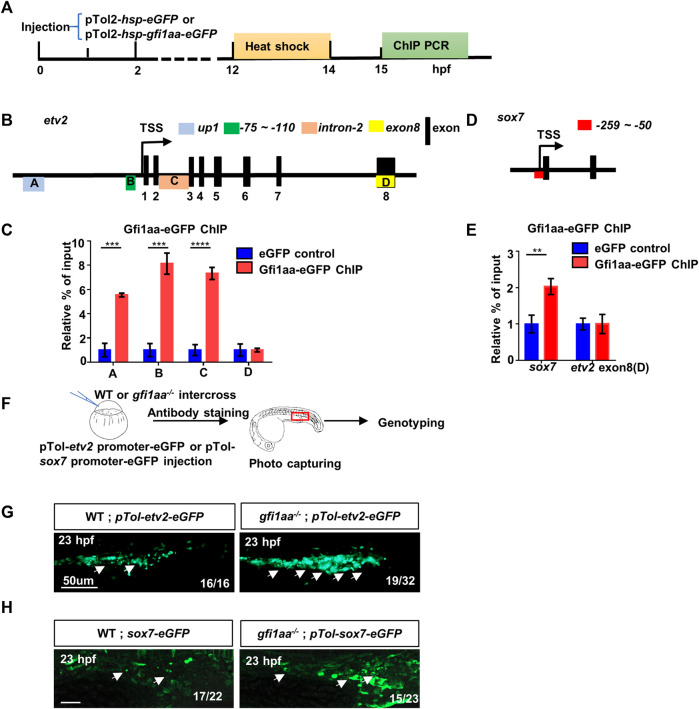
FIGURE 3.
Gfi1aa directly represses etv2 and sox7 expression (A) Workflow of Gfi1aa-eGFP ChIP-PCR assay (B) Schematic diagram of etv2 gene structure. Three regulator regions up1 (Box A, blue colored), -75 ∼ -110 bp (Box B, green colored), intron-2 (Box C, orange colored) were showed on the gene structure, black boxes indicated the exons, exon-8 (Box D, yellow colored) as the control region. Box-(A–D) represented the detected region for etv2 ChIP PCR products (C) ChIP-qPCR showed Gfi1aa enriched in etv2 regulatory regions compared to eGFP control (up1, 5.5-fold; -75 ∼ -110bp, 8.1-fold; intron-2, 7.3-fold), the results were mean ± SD and generated from three independent experiments (****p < 0.0001, ***p < 0.001, t-test) (D) Schematic diagram of sox7 gene structure. Red box represented the detected region for sox7 ChIP PCR products (E) ChIP-qPCR showed 2-fold of Gfi1aa enriched in sox7 regulatory regions compared to eGFP control. The results were mean ± SD and generated from three independent experiments (**p < 0.01, t-test) (F–H) Gfi1aa was a transcription repressor for etv2 and sox7 (F) The scheme of transient GFP reporter assay for pTol-etv2-eGFP construct and pTol-sox7-eGFP construct. The red box indicated the image region (G,H) Transient expression of pTol-etv2-eGFP construct (G) and pTol-sox7-eGFP construct (H) in WT and gfi1aa −/− mutant embryos. Fluorescence in the ICM region was monitored at 23 hpf. Scale bar: 50 μm.