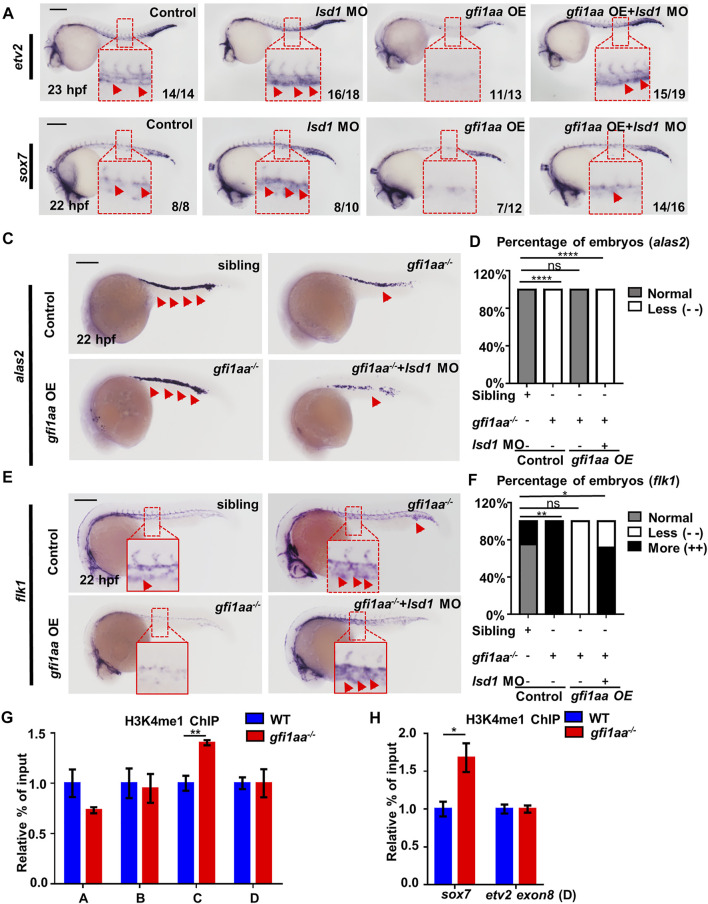
FIGURE 5.
Gfi1aa targets etv2 and sox7 in an Lsd1-dependent manner (A,B) Gfi1 repression activity on etv2 and sox7 transcription depends on Lsd1. Expression of etv2 (A) and sox7 (B) in WT, lsd1 MO, gfi1aa overexpression (gfi1aa-OE) embryos, and gfi1aa-OE embryos co-injected with 1 pmol lsd1 MO. gfi1aa-OE embryos were the progenies of hsp-gfi1aa-eGFP transgenic fish. The red boxes indicate the magnification of etv2 signals (A) and sox7 signals (B) in the ICM region. n ≥ 10 embryos for each group. The numbers in the bottom right corner indicate the percentage of embryos exhibiting the representative expression of indicated genes. Scale bar: 200 μm (C,D) Expression (C) and analysis (D) of erythroid marker alas2 in sibling, gfi1aa −/− mutant, gfi1aa-OE rescued gfi1aa −/− mutant and gfi1aa −/− mutant with gfi1aa-OE and lsd1-MO at 22 hpf (E,F) Expression (E) and analysis (F) of endothelial marker flk1 in sibling, gfi1aa −/− mutant, gfi1aa-OE rescued gfi1aa −/− mutant and gfi1aa −/− mutant with gfi1aa-OE and lsd1-MO at 22 hpf. The red boxes indicate the magnification of ICM region, and the red arrows indicate WISH signals (****p < 0.0001, **p < 0.01, *p < 0.05, ns, no significant, Fisher exact tests, n ≥ 10 embryos for each group). Scale bar: 200 μm (G,H) H3K4me1 levels at etv2 intron-2 locus and sox7 promoter were inhibited by Gfi1aa. ChIP-qPCR showed H3K4me1 level at etv2 gene loci (G) and sox7 promoter (H) in AB and gfi1aa −/− mutant embryos (The error bars represent three technical replicates and two independent experiments were performed, mean ± SEM; **p < 0.01; t-test).