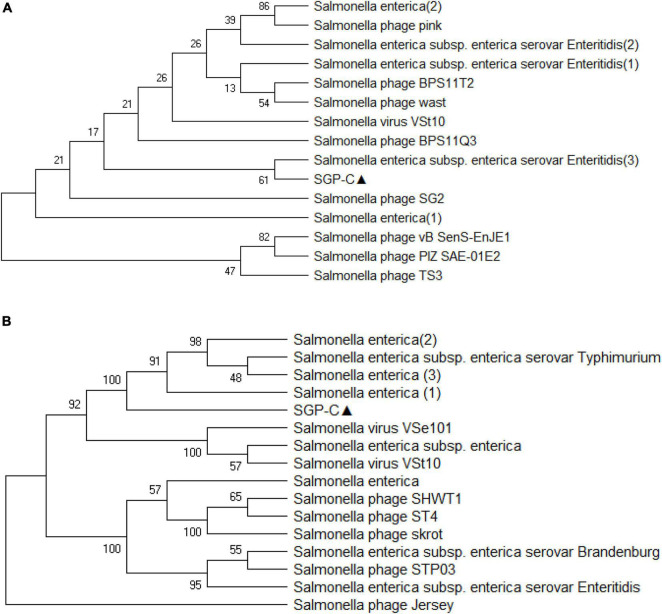
FIGURE 9.
The evolutionary history was conducted using the UPGMA method in MEGA.X based on top 15 and 16 protein homologs found on GenBank search for Tail-spike protein (A) and DNA polymerase (B). Total of 500 bootstrap replicates in which tree associated taxa clustered together are shown next to branches. Poisson correction method along with units of the number of amino acid substitute per site are used for determining evolutionary distances. There was a total of 684 positions in the first and 774 in second data set.