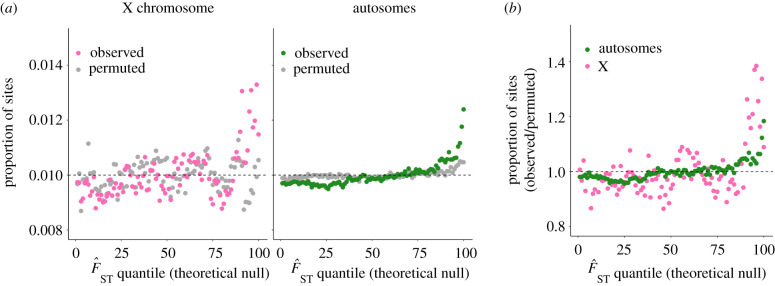
Figure 1.
Polygenic signals of SA polymorphism on the X chromosome and autosomes relative to ‘no sex-differential selection’ nulls. (a) Proportion of X-linked sites (grey, permuted; pink, observed) and autosomal sites (grey, permuted; green, observed) in each of 100 quantiles of a simulated null distribution for , with the null distributions described by equations (2.3a) and (2.3b), respectively. In the absence of sex-differential selection, approximately 1% of sites should fall into each quantile of the simulated null distribution. (b) Observed/permuted ratio of the proportion of sites in each of 100 quantiles of the simulated null distribution, for X-linked (pink) and autosomal (green) sites, respectively. Both the above panels used the full set of NX = 229 196 and NAuto. = 7 851 642 imputed sites because: (i) signals of SA polymorphism relative to null distributions cannot be artificially inflated by LD between sites (when considering X-linked and autosomal sites separately; see [48]); and (ii) power to detect X versus autosome differences is reduced by LD pruning. We present equivalent figures for LD-pruned data as electronic supplementary material, figure S7. (Online version in colour.)