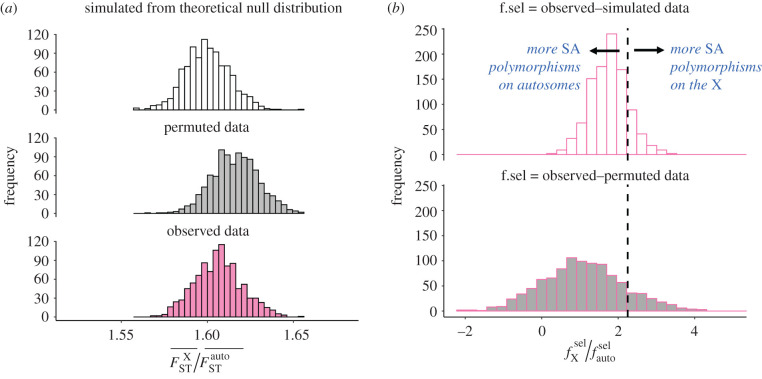
Figure 2.
Comparing polygenic signals of SA polymorphism on the X chromosome and autosomes. (a) Distribution of the ratio of mean () based on 1000 bootstrap replicates for each of our three classes of data: simulated data from the theoretical null distribution (top), permuted data (middle) and observed data (bottom), illustrating the elevation in X-linked relative to autosomal (i.e. ), even in the absence of SA polymorphism (i.e. in simulated and permuted data). (b) Distribution of the ratio of estimated X-linked to autosomal inflation in ( and ), across 1000 bootstrap replicates. The top panel uses observed and simulated data to estimate and , while the bottom panel uses observed and permuted data to estimate and . The dashed vertical line shows the theoretically predicted 9/4 X-to-autosome ratio when randomly distributed balanced SA polymorphisms account for the inflations of FST on each chromosome type. In both panels, we used a set of LD-pruned sites, rather than the full data, to avoid biases arising from differences in the extent of hitchhiking between autosomes and the X (see Methods). (Online version in colour.)