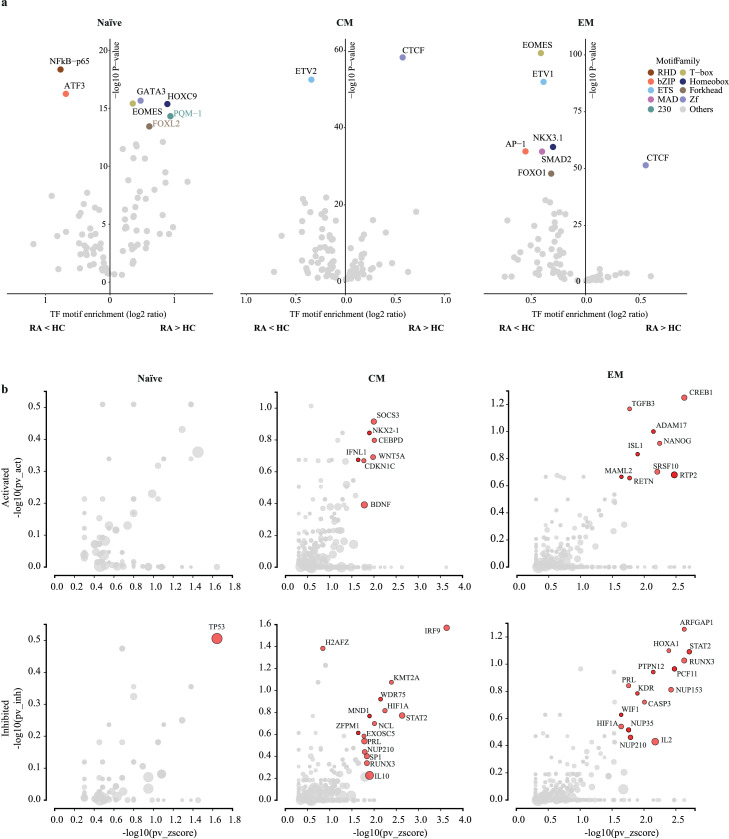
Figure 3.
Upstream regulators of differentially accessible regulatory sites in RA CD4 T cells. (a) Identification of transcription factor (TF) motifs enriched at sites more (right) or less open (left) in RA for each T cell subset. For each motif family, only the top ranked TF is shown. X-axis denotes enrichment of each motif over background, Y-axis the enrichment log10 p-value as reported by HOMER. (b) Upstream regulator genes predicted to be activated (top) or inhibited (bottom) in RA compared to HC for each T cell subset. Y-axis compares the number of targets consonant with the prediction to the total number of targets. X-axis denotes the overrepresentation of these targets compared to the number of target genes expected just by chance. Upstream regulators with a significant combined p-value < 0.05 are labelled (red) as reported by iPathwayGuide.