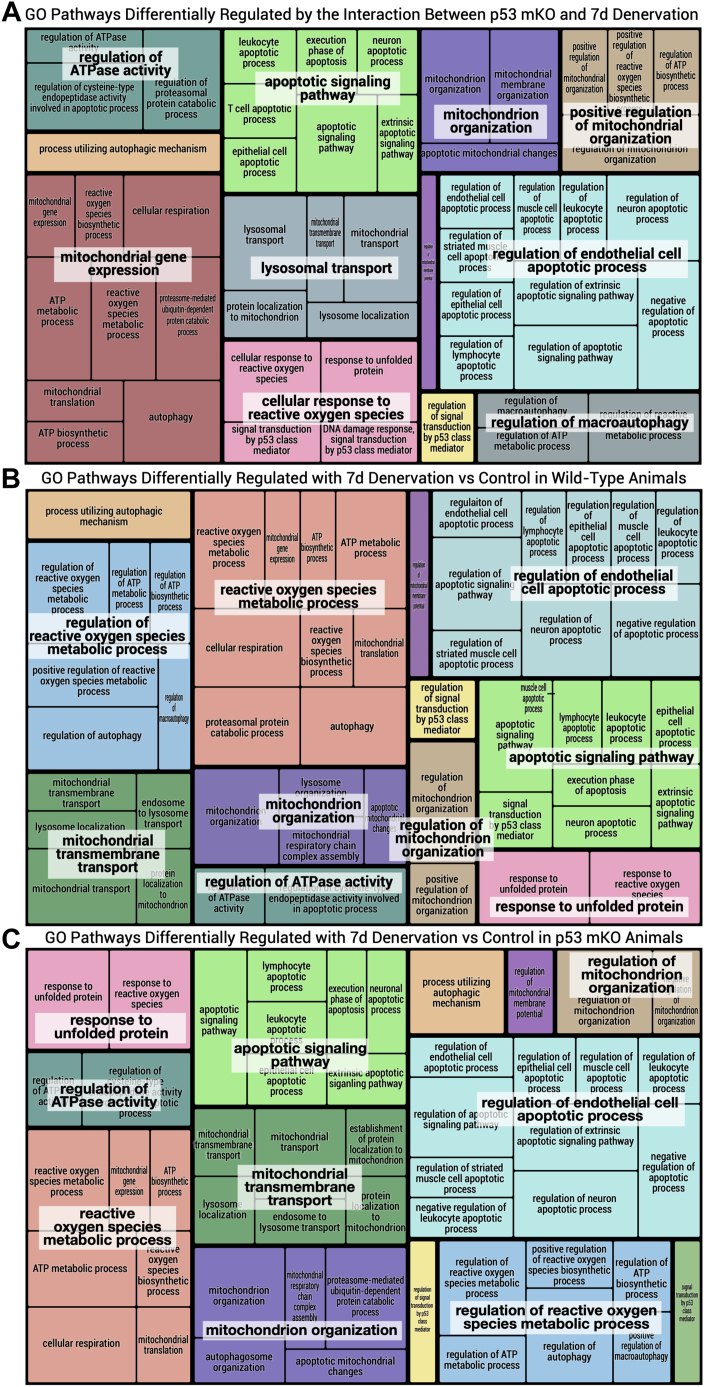
Figure 3.
REVIGO (reduce and visualize GO) tree map of GO terms elevated in response to denervation. GO terms pertaining to mitochondrial quality control were selected and used to generate tree maps based on their p value as described previously using the REVIGO web-based application (24). Selection terms for GO pathways included “mitochondria,” “autophagy,” “mitophagy,” “lysosome,” “ROS” or “reactive oxygen species,” “UPR” or “unfolded protein response,” “apoptosis,” “p53,” “protein synthesis,” and “degradation” were used to filter GO terms in order to represent the unique pattern of differentially expressed gene sets in WT versus p53 mKO animals with 7 days of hindlimb denervation. A, tree map representing the GO pathways that are differentially regulated by denervation in p53 mKO as compared with WT animals (based on the analysis of the interaction of genotype and denervation). B and C, individual tree maps of GO pathways that are differentially regulated by chronic disuse in WT animals and p53 mKO animals, respectively. GO, Gene Ontology; mKO, muscle-specific KO.