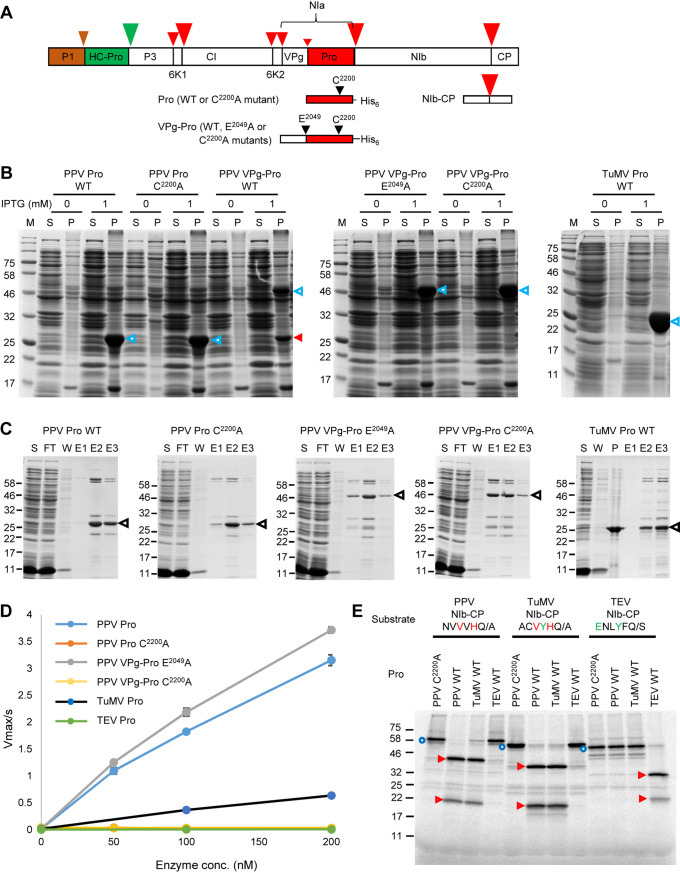
FIG 1.
Activity and specificity of purified recombinant potyvirus proteases. (A) Schematic representation of the PPV polyprotein. Autoproteolytic cleavage sites that are processed by the P1 and HC-Pro proteases are indicated with brown and green arrowheads, respectively. Cleavage sites that are processed by the NIa protease are marked with red arrowheads. The size of the arrowhead represents the relative cleavage efficiency at each site. Portions of the polyprotein used in this study are shown, including the Pro and VPg-Pro used for production of recombinant proteases and the NIb-CP partial polyprotein used as a substrate for the proteases. The position of the catalytic triad cysteine (C2200, numbering from the beginning of the PPV polyprotein) and of the glutamic acid (E2049) at the P1 position of the VPg-Pro cleavage site are also shown. (B) Expression of various forms of the PPV and TuMV NIa proteases. Protein expression was induced by the addition of 1 mM IPTG, and protein extracts were run on SDS-PAGE after separation into supernatant fractions (S) containing soluble proteins and pellet fractions (P) containing insoluble proteins. Control lanes (0 mM IPTG) were included that corresponded to S and P fractions from Escherichia coli grown in the absence of IPTG. Migration of the full-length expressed proteins is shown with the blue arrowheads. The mature Pro released after autocatalytic cleavage of the PPV VPg-Pro WT is indicated by the red arrowhead. Please note that this fragment comigrates with the mature Pro expressed from the PPV Pro wild type (WT) and PPV Pro C2200A constructs (calculated molecular mass, 28.9 kDa). M, protein ladder with the molecular mass (kDa) indicated on the left. (C) Purification of PPV and TuMV proteases. SDS-PAGE gels showing the purification of recombinant proteases. Proteases were partially purified from the soluble protein fraction using the Ni-NTA resin column. Expected migration of the expressed proteases is indicated by the black arrowheads. Other bands are likely minor contaminant E. coli proteins. FT, flowthrough from the Ni-NTA resin column; W, wash collections; E, elution fractions; S, supernatant fraction containing soluble proteins; P, pellet fraction containing insoluble proteins. The migration of molecular mass markers is indicated on the left. (D) Activity of purified potyvirus proteases tested using the MCA-QA-DNP fluorescent peptide substrate, which is derived from the PPV NIb-CP cleavage site. Reactions were conducted in the assay buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.1% Brij35). The substrate concentration was 5 μM, and the enzyme concentration ranged from 50 to 200 nM. Values are reported as the mean ± standard deviation (SD) from triplicate measurements. (E) In vitro processing of partial viral polyproteins (PPV, TuMV, or TEV NIb-CP) by the PPV, TuMV, and TEV Pro[s]. NIb-CP from PPV, TuMV, and TEV were synthesized in vitro using rabbit reticulocytes extracts. Translation products were then incubated at 16°C for 16 h in the presence of 50 nM wild-type or mutated PPV, TuMV, or TEV protease (as indicated above each lane). The full-length precursor protein and cleavage products were separated by 12% SDS-PAGE. The migration positions of molecular mass markers are indicated on the left. Full-length substrate proteins are indicated with blue circles, and cleaved fragments are indicated with red arrowheads.