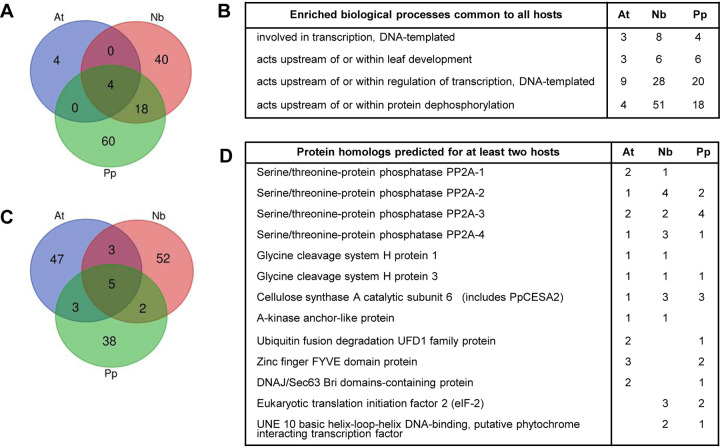
FIG 2.
Analysis of putative protein targets in natural or experimental hosts of PPV. (A) Venn diagram showing shared enriched biological processes associated with putative target proteins in A. thaliana (At), N. benthamiana (Nb), and P. persica (Pp). Gene ontologies (GOs) associated with identified protein targets were determined using the TAIR database annotation for the closest homolog as determined by BLASTP. For each host, GOs with adjusted P values over 0.05, as well as GOs corresponding to fewer than three proteins, were eliminated prior to the comparison. See Excel Sheet S1 in the supplemental material for additional information. (B) List of enriched GOs (biological process) shared in all three plant hosts. The number of proteins associated with each GO is indicated for each plant host. (C) Venn diagram showing common putative protein targets among the three plant hosts. For N. benthamiana and P. persica, BLASTP was used to identify the most related A. thaliana gene. The lists of corresponding A. thaliana genes generated for each host were then compared. See Excel Sheet S1 for additional information. (D) List of common target proteins. The numbers of protein isoforms identified for each common gene and for each host are indicated.