FIG 6.
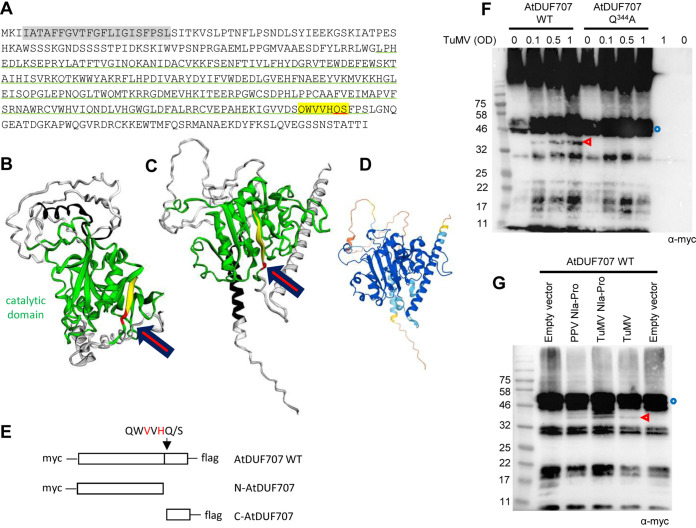
Partial in vivo cleavage of AtDUF707. (A) Sequence of AtDUF707. The predicted catalytic domain is underlined in green. A strongly predicted trans-membrane helix is highlighted in gray. Letters highlighted in yellow correspond to the P6 to P1′ positions of the predicted cleavage site with the P1 and P1′ positions underlined in red. (B to D) Structural models of the AtDUF707 protein predicted using Phyre2 (see Materials and Methods) (B) or AlphaFold (PDB file of the model available at https://www.alphafold.ebi.ac.uk/) (C). For both models, the color scheme is as follows: catalytic domain in green, putative trans-membrane helix in black, P6 to P2 positions of the predicted cleavage site in yellow, and P1 to P1′ position of the cleavage site in red. The position of the cleavage site is indicated by the red arrow. The degree of confidence in the Phyre2 model (B) differed by domain of the protein. The predicted catalytic domain was modeled with a high degree of confidence (90.4%), based in part on the solved structure of the catalytic domain from ATP synthase subunits of orf6 of the f1-atpase2 operon of Rhodobacter blasticus (PDB ID 2QGI) and on those of several other transferase domains from acetylgalactosaminyltransferases. Other regions of the protein were not modeled with a high degree of confidence. The degree of confidence in the AlphaFold model (C) also differed by region of the polyprotein (color scheme as in Fig. 4D) (D). In both models, the cleavage site is located at the base of a β-sheet, which is part of the predicted catalytic domain. (E) Schematic representation of AtDUF707 protein fused to the Myc and Flag epitopes. The predicted cleavage site is shown by the black arrow. Cleavage by the NIa protease would release the Myc-tagged N-terminal fragment (N-AtDUF707) and the Flag-tagged C-terminal fragment (C-AtDUF707). (F) Cleavage of AtDUF707 by the NIa protease released from the TuMV polyprotein. N. benthamiana leaves were agroinfiltrated with a mixture of A. tumefaciens transformed with the AtDUF707 constructs (WT or Q344A mutant) (OD = 1.0) and A. tumefaciens transformed with the TuMV infectious clone (OD = 0.1 to 1, as indicated above each lane). Control experiments did not incorporate A. tumefaciens transformed with the TuMV infectious clone (marked as OD = 0). (G) Cleavage of DUF707 by PPV or TuMV Pro[s]. N. benthamiana leaves were agroinfiltrated with a mixture of A. tumefaciens transformed with the AtDUF707 WT construct (OD = 0.5) and A. tumefaciens transformed with the empty vector (OD = 0.1), PPV Pro construct (OD = 0.1), TuMV Pro construct (OD = 0.1), or TuMV infectious clone (OD = 1). (F and G) Plant protein extracts were separated by 12% SDS-PAGE and transferred to PVDF membranes. Immunoblots were probed with anti-Myc antibodies. Blue circles and red arrowheads indicate the full-length protein and cleavage products, respectively. The large bands at the top of the gel in panel F are likely due to aggregation of the protein.