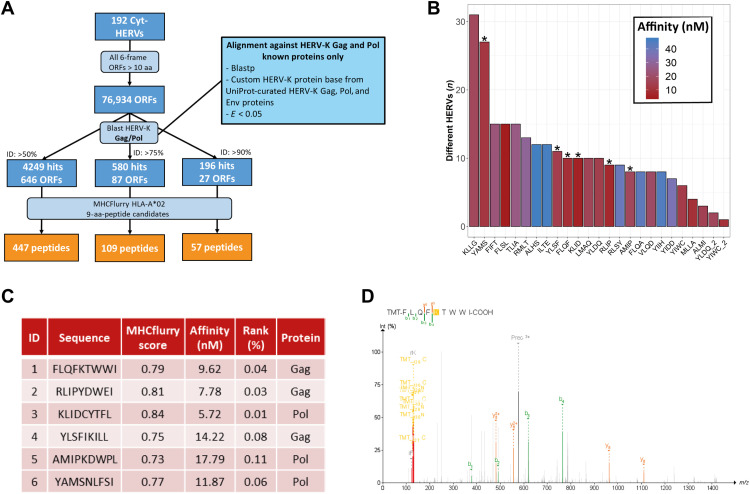
Fig. 2. Selection of shared HLA-A2 epitopes derived from Gag and Pol HERV-K/HML-2.
(A) Flow chart of peptide selection from cyt-HERV sequences. aa, amino acid. (B) Bar chart of the top 25 most shared peptides predicted as strong HLA-A*02 binders among the 192 cyt-HERVs. Selected peptides (P1 to P6) are marked with a star. (C) Characteristics of the six selected HLA-A2 epitopes. Nine-mer peptides were selected according to their predicted HLA-A*02 affinity (considering strong binders for percentile ranks of ≤0.5) and the number of HERVs containing their sequences. (D) MS/MS detection of P1 epitope in a sample from the CPTAC breast cancer prospective dataset. MS/MS spectrum is identified by Pepquery analysis (Peptide Spectrum Match, P value of 0.0063). m/z, mass/charge ratio.