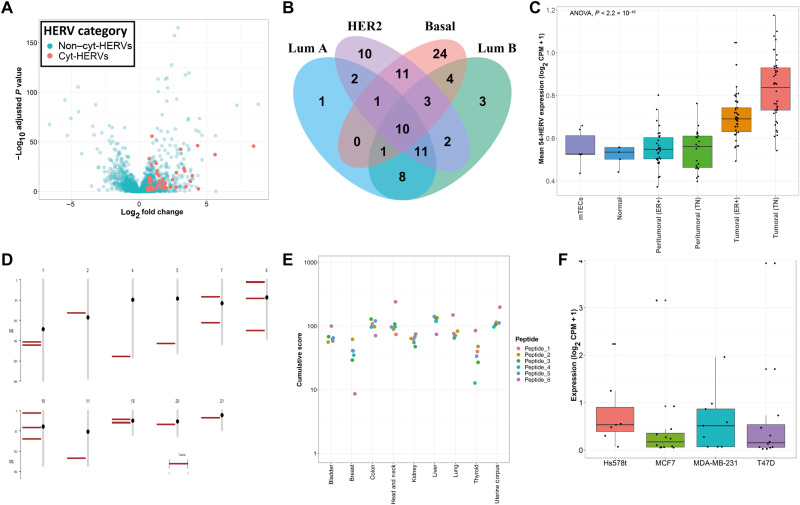
Fig. 3. Shared CD8+ T cell epitopes derived from conserved Gag and Pol HERV-K/HML-2 motifs are expressed in TNBC.
(A) Volcano plot of HERVs overexpressed in basal breast cancer subtype compared to peritumoral samples in TCGA database. cyt-HERVs are represented in red. (B) Venn diagram of the total number of cyt-HERVs overexpressed in each subtype of breast cancer in TCGA database. (C) Mean expression of the 54 cyt-HERVs overexpressed in TCGA basal subtype, in the independent database of Varley et al. (25), and in mTECs. (D) Chromosomal location of the 18 peptide-containing HERVs overexpressed in the basal breast cancer subtype. Each bar corresponds to an HERV locus. (E) Cumulative expression score of the epitope-containing HERVs in TNBC versus normal tissues. This score was calculated for each epitope and each available peritumoral sample. (F) Expression of the 18 peptide-containing CAHs in the breast cancer basal (Hs578t and MDA-MB-231) and luminal A (MCF7 and T47D) cell lines analyzed by riboseq.