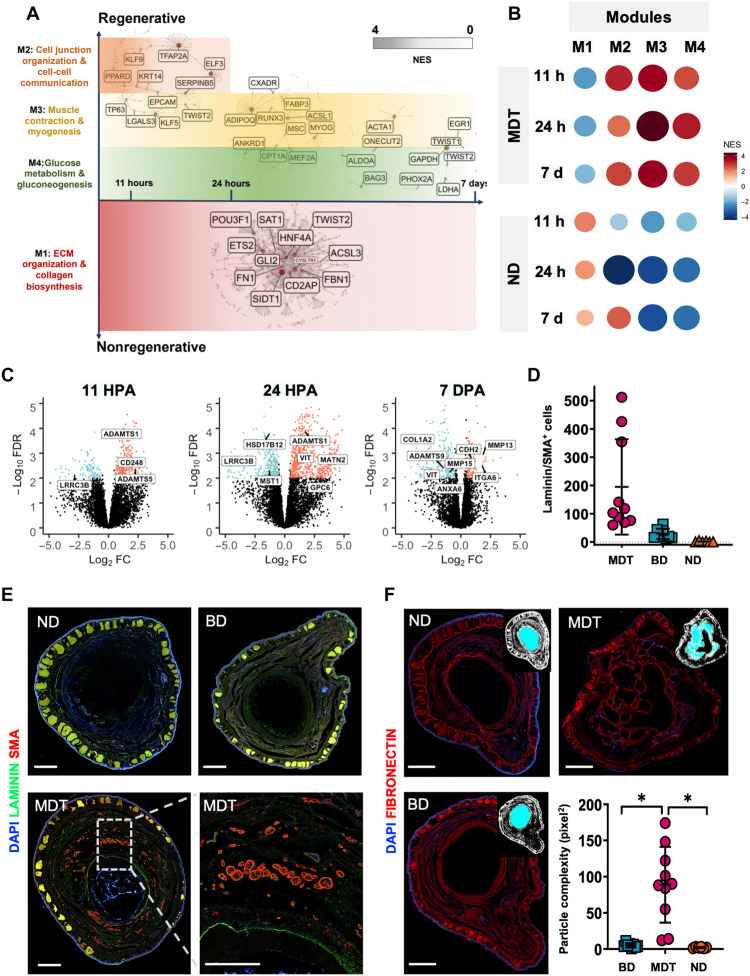
Fig. 5. GSEA revealed four distinct expression modules indicative of complex tissue repatterning.
CEMiTool was used to identify covarying gene sets as a function of high fold changes in the MDT (CT) and ND animals, yielding modules M1 to M4. (A) Relative expression of modules between time points and conditions (dark red circles = up-regulation; dark blue circles = down-regulation). (B) M2 to M4 were up-regulated in MDT animals versus ND across all time points, peaking at 24 hpa; however, M1 was down-regulated in the MDT condition, most highly at 11 hpa, and decreasing to 7 dpa. M3 genes were up-regulated in MDT relative to ND including acetylcholine activity and muscle activation. M4 genes were up-regulated in MDT relative to ND including glucose metabolism. (C) GO data within the ECM development pathway indicated up-regulation of ADAM family genes at 11 and 24 hpa; however, they were down-regulated at 7 dpa. (D and E) Cross-sectioned regenerates (18 mpa) were double fluorescent–stained for markers of vascularization: smooth muscle actin (red) and basement membrane (green). SMA/laminin+ bundles were increased in MDT animals relative to device only or ND groups (P < 0.001). (F) Fibronectin+ cartilaginous cores of regenerates displayed increased structural complexity associated with the MDT group relative to all other groups (*P < 0.05). Scale bars, 1 mm. Means and SD are presented.