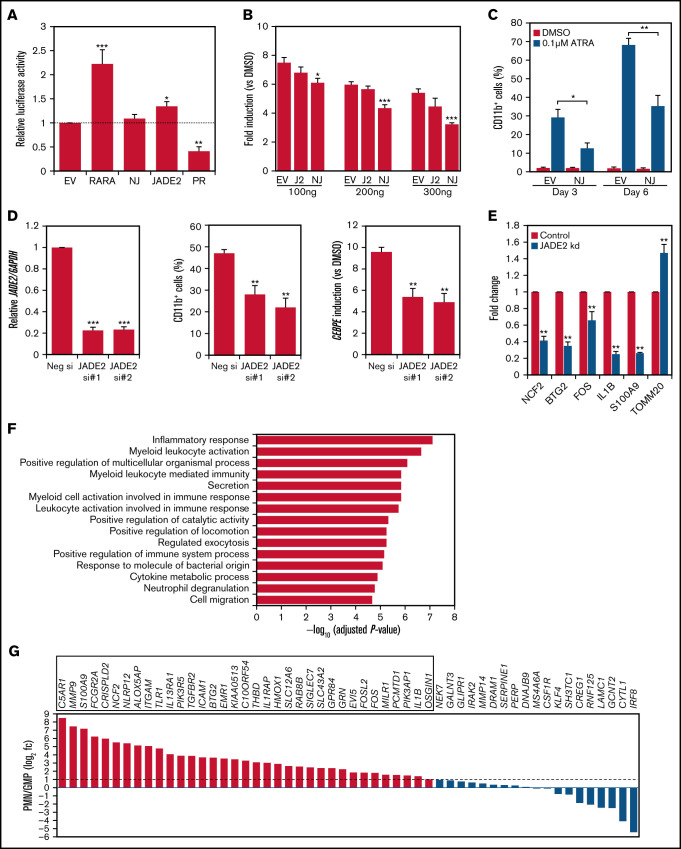
Figure 2.
Molecular characterization of NUP98-JADE2 and JADE2 in ATRA-mediated responses and myeloid differentiation. (A) HeLa cells were cotransfected with RARE reporter and the indicated pCMV-HA plasmids. EV, empty vector; NJ, NUP98-JADE2; PR, PML-RARA. Luciferase activities were determined as described in supplemental Methods. (B) HeLa cells were cotransfected with RARE reporter and an increasing pCMV-HA-NUP98-JADE2 (NJ), pCMV-HA-JADE2 (J2), or the empty pCMV-HA (EV), and then treated with ATRA or DMSO for 6 hours before analysis. Results are presented as fold induction vs DMSO treatment. (A-B) *P < .05, **P < .01, and ***P < .001 vs EV, respectively by 1-way analysis of variance followed by Dunnett test. (C) NB4 cells transduced with NUP98-JADE2 (NJ) or control (EV) lentiviruses were treated with ATRA or DMSO. CD11b on GFP-positive cells was measured by flow cytometry. *P < .05 and **P < .01, respectively by paired t test. (D) Left, confirmation of JADE2 knockdown in NB4 cells after 24 hours of siRNA transfection by quantitative RT-PCR. After the transfection, cells were treated with 0.1 µM of ATRA or DMSO for 3 days. CD11b was measured by flow cytometry (middle) and CEBPE by quantitative RT-PCR and normalized to GAPDH (right). **P < .01 and ***P < .001 vs the negative siRNA, respectively by 1-way analysis of variance followed by Dunnett test. (E) Quantitative RT-PCR to validate selected JADE2 target genes in NB4 cells after 48 hours of siRNA transfection. **P < .01 vs negative siRNA by the Mann-Whitney U test. (A-E) Results are expressed as mean plus standard error from 3 independent experiments. (F) Pathway analysis of the 56 downregulated genes by ConsensusPathDB.15 The top 15 enriched Gene Ontology Biological Processes are shown. (G) Expression of the downregulated genes in purified polymorphonuclear cells (PMN) and granulocyte-monocyte progenitors (GMP). Data (from GSE42519) were available for 54 of the downregulated genes. Genes that are upregulated by ≥twofold (log2 fc≥1) in PMN are boxed.