Summary
Soil availability of inorganic ortho-phosphate (PO43−, Pi) is a key determinant of plant growth and fitness.1 Plants regulate the capacity of their roots to take up inorganic phosphate by adapting the abundance of H+-coupled phosphate transporters of the PHOSPHATE TRANSPORTER 1 (PHT1) family2 at the plasma membrane (PM) through transcriptional and post-translational changes driven by the genetic network of the phosphate starvation response (PSR).3, 4, 5, 6, 7, 8 Increasing evidence also shows that plants integrate immune responses to alleviate phosphate starvation stress through the association with beneficial microbes.9, 10, 11 Whether and how such phosphate transport is regulated upon activation of immune responses is yet uncharacterized. To address this question, we first developed quantitative assays based on changes in the electrical PM potential to measure active Pi transport in roots in real time. By inserting micro-electrodes into bulging root hairs, we were able to determine key characteristics of phosphate transport in intact Arabidopsis thaliana (hereafter Arabidopsis) seedlings. The fast Pi-induced depolarization observed was dependent on the activity of the major phosphate transporter PHT1;4. Notably, we observed that this PHT1;4-mediated phosphate uptake is repressed upon activation of pattern-triggered immunity. This inhibition depended on the receptor-like cytoplasmic kinases BOTRYTIS-INDUCED KINASE 1 (BIK1) and PBS1-LIKE KINASE 1 (PBL1), which both phosphorylated PHT1;4. As a corollary to this negative regulation of phosphate transport by immune signaling, we found that PHT1;4-mediated phosphate uptake normally negatively regulates anti-bacterial immunity in roots. Collectively, our results reveal a mechanism linking plant immunity and phosphate homeostasis, with BIK1/PBL1 providing a molecular integration point between these two important pathways.
Keywords: plant immunity, kinase, phosphate, PHT1, root microbiome, electrophysiology
Highlights
-
•
Real-time, PHT1-mediated Pi transport can be measured in roots via PM depolarization
-
•
Immune activation represses Pi transport in a BIK1/PBL1-dependent manner
-
•
PHT1s are direct substrates of BIK1 and PBL1
-
•
Pi uptake modulates anti-bacterial immunity and the root microbiome
Phosphate (Pi) is a crucial determinant of plant growth and fitness. Dindas, DeFalco, et al. show that immune activation in roots actively represses Pi uptake. This inhibition is achieved via direct phosphorylation of key Pi transporters by the central immune kinases BIK1 and PBL1, which leads to increased anti-bacterial defense.
Results and discussion
Pi-induced membrane depolarization as a real-time readout of PHT1 activity
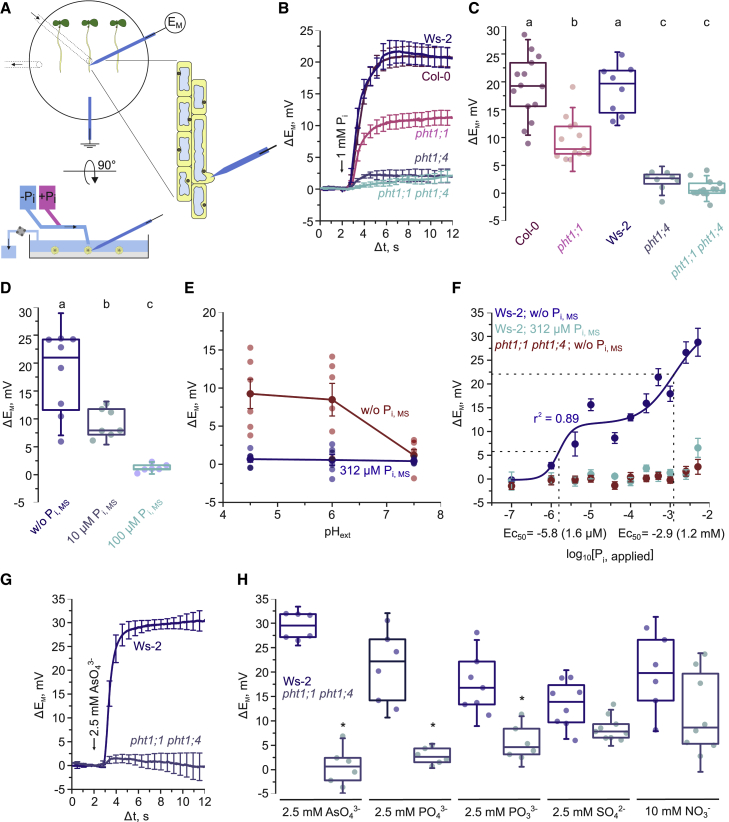
PHT1s act as H+-coupled transporters that utilize the proton motive force across the plasma membrane (PM) to transport the inorganic ortho-phosphate (Pi) anion into the cytosol.2 Such electrogenic processes can be directly monitored in planta and in real time through changes in the electrical PM potential via intracellular micro-electrodes.12, 13, 14 To test whether external Pi application results in a change of the Arabidopsis root PM potential, we impaled micro-electrodes in the tips of bulging root hair cells of Pi-starved 5-day-old seedlings (Figure 1A). While the resting potential of root hair cells was on average ∼−160 mV (Figure S1A), Col-0 and Ws-2 wild-type root hair cells rapidly depolarized to amplitudes between 10 and 30 mV within seconds after Pi application (Figures 1B and 1C), suggesting electrogenic Pi uptake.
Figure 1.
Application of Pi induces a depolarization of the root hair PM potential representative of PHT1;4-mediated H+/PO43− transport
(A) Schematic of the experimental setup of micro-electrode-based Pi transport measurements in intact Arabidopsis seedlings.
(B) Averaged responses of the PM potential to application of 1 mM Pi at pH 6 on wild-type and pht1 mutant seedlings. Curves are normalized and aligned to the point of Pi application (arrow, Δt = 2 s). Error bars show ± SEM (n = 8–14).
(C) ΔEM values averaged over Δt = 4.5 s to 5.5 s from curves in (B).
(D) ΔEM values in response to application of 1 mM Pi at pH 6. Col-0 plants were grown on media supplemented without or with the indicated Pi concentrations.
(E) Straight lines connect average ΔEM values in response to application of 1 mM Pi for external pH values of 4.5, 6, and 7.5 and Col-0 seedlings grown on media supplemented without or with 312 μM Pi. Error bars show ± SEM.
(F) Dose-response curve of Pi-induced root hair depolarization. The solid line represents a bi-phasic dose-response curve. The dashed lines indicate the half maximal effective concentration (EC50) values. Error bars show ± SEM (n = 5–8).
(G) Averaged responses of the PM potential to application of 2.5 mM arsenate at pH 6. Curves are normalized and aligned to the point of Pi application (Δt = 2). Error bars show ± SEM (n = 6).
(H) ΔEM values at Δt = 6.5–7.5 from curves in (G) and from Figure S1G for other ions. Stars at the top of the panel indicate significance between pht1;1 pht;1,4 and Ws-2.
Boxes indicate the 25th and 75th percentile, the horizontal line shows the median, and error bars represent ± SD. Closed circles show individual measurements. When indicated, significance was tested with a one-way ANOVA and a post hoc Tukey test. Equal letters at the top of the panel indicate p > 0.05. See also Figure S1.
Among the 9 PHT1s encoded by the Arabidopsis genome, PHT1;1 and PHT1;4 were previously identified to account for the majority of total Pi uptake based on radioactive assays in whole seedlings.3,4 To test whether this Pi-induced depolarization is mediated by PHT1s, we impaled root hair cells of pht1;1 and pht1;4 mutants. The pht1;1 single mutant (in Col-0 background) showed ∼50% reduction of the average response, while both the pht1;4 (in Ws-2 background) and the pht1;1 pht1;4 (Ws-2) mutants showed no such depolarization at all (Figures 1B and 1C). We confirmed the phenotype of pht1;4 in two additional homozygous knockout alleles in the Col-0 ecotype (Figures S1B–S1D).
In addition to local mechanisms to cope with low Pi,5,15 which result in a reshaping of the root system architecture to promote top-soil foraging by inhibiting cell proliferation and expansion in the meristematic and elongation zone of the primary root,16 metabolized Pi in the form of inositol polyphosphates (InsPs) is perceived at the systemic level by SPX-domain-containing proteins.17, 18, 19, 20, 21, 22, 23 Low Pi levels trigger the phosphate starvation response (PSR), which favors root over shoot development and vacuolar release of sequestered Pi24,25 as well as an increase in Pi uptake capacity via accumulation of PHT1s at the PM. The accumulation of PHT1s at the root cell PM under deficient Pi conditions is driven by transcriptional upregulation,3,4 a C-terminal phospho-switch regulating transit from the endoplasmic reticulum to the PM,6 and inhibition of PHT1 degradation.6,7 Consistent with an increased accumulation of PHT1s at the PM upon Pi starvation, supplementing the growth medium with greater than 10 μM Pi suppressed Pi-induced depolarizations with no effect on the resting potential (Figures 1D and S1E). Combined, these results suggest that the Pi-induced depolarization requires PHT1s, with PHT1;4 as the dominant contributor on the timescale and in the tissues of our experiments.
To further corroborate whether the observed Pi-induced depolarization is indeed a direct real-time readout of H+-coupled Pi uptake, we tested its pH and Pi dependence. Challenging seedlings with solutions buffered to different pH values revealed a strong dependence of the Pi-induced depolarization on an acidic pH and thus an inward-directed H+ gradient (Figure 1E), despite the counteracting acidification effect exerted on the PM resting potential (Figure S1F). A bi-phasic logistic function was fitted to the dose-response relation of applied Pi, suggesting apparent half-maximal responses at 1.6 μM and 1.2 mM Pi (Figure 1F), which are in agreement with values reported previously for in planta Pi-transport kinetics.2,13,26,27
The clear absence of a Pi-induced depolarization in pht1;1 pht1;4 root hair cells (Figures 1B and 1F) provided us with a tool to test the selectivity of PHT1s in vivo. Challenging the roots of Pi-starved Ws-2 and pht1;1 pht1;4 seedlings with different potential anionic PHT1 substrates suggested a selectivity order of AsO43− ≥ PO43− > PO33−, in keeping with the known permeability of PHT1s to arsenate,3 although the depolarization induced by SO42− or a high concentration of NO3− did not depend on the presence of PHT1;1 and PHT1;4 (Figures 1G, 1H, and S1G). Taken together, our in vivo electrophysiological analyses show that the rapid depolarization induced by external Pi application is an unequivocal real-time representation of the mH+/nPO43− (with m/n > 3) transport activity of PHT1;4 and, to lesser extent, PHT1;1.
Immune signaling inhibits PHT1-mediated Pi transport
Beyond physiological adaptions, plants have also developed beneficial associations with root-colonizing microbes, such as arbuscular mycorrhiza (AM) fungi, to alleviate Pi limitation in most natural soils.28 It was, however, unclear how plant defense responses and nutritional cues are integrated to allow colonization with beneficial microbial communities. Recent work uncovered that the AM non-host Arabidopsis represses defense responses in correlation with Pi stress to facilitate colonization by beneficial microbes that assist with Pi uptake,9,29 while in rice, a PHOSPHATE STARVATION RESPONSE (PHR)-centered transcriptional network regulates AM symbiosis.11 Importantly, the PHR1-dependent genetic network coordinating the PSR influences the assembly of the root-associated microbiome by direct transcriptional regulation of immunity-related genes, suggesting a PHR1-mediated link between the PSR and plant immunity.9,10 The plant immune system is traditionally classified into pattern-triggered immunity (PTI) and effector-triggered immunity (ETI).30 PTI defines the first layer of defense responses, because its molecular signaling pathways are activated through the perception of microbe-associated molecular patterns (MAMPs), pathogen-associated molecular patterns (PAMPs), or damage-associated molecular patterns (DAMPs) by cell-surface-localized pattern-recognition receptor (PRR) complexes.31 It remained, however, unresolved if a direct integration of PTI into Pi homeostasis exists.
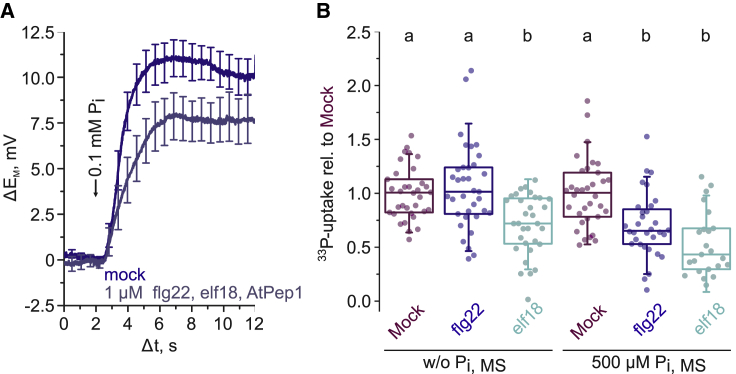
Having established a real-time readout for PHT1;4-mediated Pi uptake, we next tested whether activation of PTI has an effect on Pi uptake. To do so, we pre-treated Arabidopsis seedlings with a cocktail of molecular patterns that induce PTI (flg22, elf18, and AtPep1) prior to root hair impalement. This pre-treatment led to a significant reduction of the Pi-induced depolarization in Pi-starved Col-0 root hair cells (Figures 2A, S2A, and S2B). We corroborated this result with radioactive Pi assays that probe Pi uptake in Arabidopsis seedling roots (Figures 2B and S2C). These experiments revealed a flg22- and elf18-elicited reduction in the Pi uptake rate when seedlings were grown under sufficient Pi conditions, while, in Pi-starved seedlings, only elf18 led to a significant reduction of the Pi uptake rate. Together, these results suggest that activation of PTI signaling negatively regulates PHT1-mediated Pi transport across the PM.
Figure 2.
Elicitor treatment attenuates PHT1;4-mediated Pi uptake
(A) Averaged responses of the PM potential to application of 0.1 mM Pi in Col-0 root hairs grown in the absence of Pi. Curves are normalized and aligned to the point of Pi application (Δt = 2). Error bars show ± SEM (n = 5–8).
(B) Boxplot of radioactive Pi uptake normalized to the respective mock control of each replicate. Boxes indicate the 25th and 75th percentile, the horizontal line shows the median, and error bars represent ± SD. Closed circles show individual measurements.
Significance was tested with a one-way ANOVA and a post hoc Tukey test. Equal letters at the top of the panel indicate p > 0.05. See also Figure S2.
RLCKs regulate PHT1-mediated Pi transport
Receptor-like cytoplasmic kinases (RLCKs) are the central executors of molecular signaling downstream of PAMP or DAMP perception by their cognate receptor complexes.32 The RLCK-VII isoform BOTRYTIS-INDUCED KINASE 1 (BIK1), along with its close homolog PBS1-LIKE KINASE 1 (PBL1), are key signaling components that act downstream of multiple PRRs, such as FLAGELLIN-SENSING 2 (FLS2), EF-TU RECEPTOR (EFR), and PEP RECEPTOR 1 and 2 (PEPR1/2), which perceive flg22, elf18, and AtPep1, respectively.33, 34, 35, 36, 37, 38 BIK1/PBL1 directly phosphorylate target proteins to regulate a number of PTI outputs, such as production of reactive oxygen species, calcium influx, and stomatal closure.37, 38, 39, 40 Notably, BIK1 was previously suggested to be a negative genetic regulator of Pi homeostasis, as bik1 plants show a decreased abundance of central PSR gene transcripts, including PHR1 and PHT1;4 and, interestingly, an increased accumulation of Pi.41 However, the molecular mechanism defining the role of BIK1 in Pi homeostasis and its potential connection to PTI remained unresolved. We thus hypothesized that direct interaction between BIK1/PBL1 and PHT1s could provide a mechanistic link between PTI and Pi homeostasis. In agreement with such a regulatory role, we observed association of BIK1-hemagglutinin (HA) with PHT1;4-GFP and FLS2-GFP, but not with the PM-marker Lti6b-GFP, via immunoprecipitation after co-expression in Nicotiana benthamiana (Figure 3A). Similarly, we observed association of BIK1-HA and PHT1;4-GFP when both were expressed under their native promoters in stable Arabidopsis lines, while BIK1-HA did not interact with Lti6b-GFP in the same assay (Figure 3B).
Figure 3.
BIK1 phosphorylates PHT1;4 and inhibits PHT1;4-mediated Pi transport
(A and B) Co-immunoprecipitation of BIK1-HA (A) after transient co-expression with PHT1;4-GFP in N. benthamiana leaves or (B) in stable transgenic Arabidopsis seedlings expressing BIK1pro:BIK1-HA with 35Spro:Lti6b-GFP or PHT1;4pro:PHT1;4-GFP. LTI6b-GFP and FLS2-GFP serve as negative and positive controls for BIK1 association, respectively. Co-immunoprecipitation was repeated three times with similar results.
(C and D) In vitro radioactive kinase assays with the indicated combinations of purified recombinant proteins. Asterisk indicates kinase-dead variant. The assays were repeated at least twice with similar results.
(E) Averaged responses of the PM potential to application of 0.1 mM Pi at pH 6 without or after a 15–30 min pre-treatment with an elicitor mix of 1 μM flg22, elf18, and AtPep1. Curves are normalized and aligned to the point of Pi application (Δt = 2). Combined data of three independent biological replicates (Figure S6) are shown. Error bars show ± SEM (n = 66–77).
(F) Normalized ΔEM values at Δt = 5.5–6.5 from curves in (E).
See also Figures S3 and S4.
We next tested whether BIK1 and PBL1 phosphorylate the large cytosolic loop or C terminus of PHT1;4 (Figure S3A) in vitro. Indeed, maltose-binding protein (MBP)-fused BIK1 and PBL1, but not their kinase-dead variants, phosphorylated the glutathione S-transferase (GST)-fused cytosolic loop of PHT1;4, but not the PHT1;4 C terminus (Figures 3C and 3D). In keeping with the high similarity and shared roles of PHT1;4 and PHT1;1, BIK1 and PBL1 also phosphorylated the cytosolic loop of PHT1;1 (Figures S3B and S3C), indicating that these PHT1s serve as BIK1 and PBL1 substrates. We investigated next whether BIK1 and PBL1 are genetically required for pattern-triggered attenuation of Pi uptake. Pre-treatment with the pattern mix did not affect the resting potential of Col-0 or bik1 pbl1 (Figure S4); however, it significantly reduced PHT1;4-mediated phosphate transport (as measured by membrane depolarization) in Pi-starved Col-0, but not bik1 pbl1 roots (Figures 3E, 3F, and S4). These results indicate that PTI signaling actively represses Pi transport in a BIK1/PBL1-dependent manner.
Inhibition of Pi transport shapes immune responses
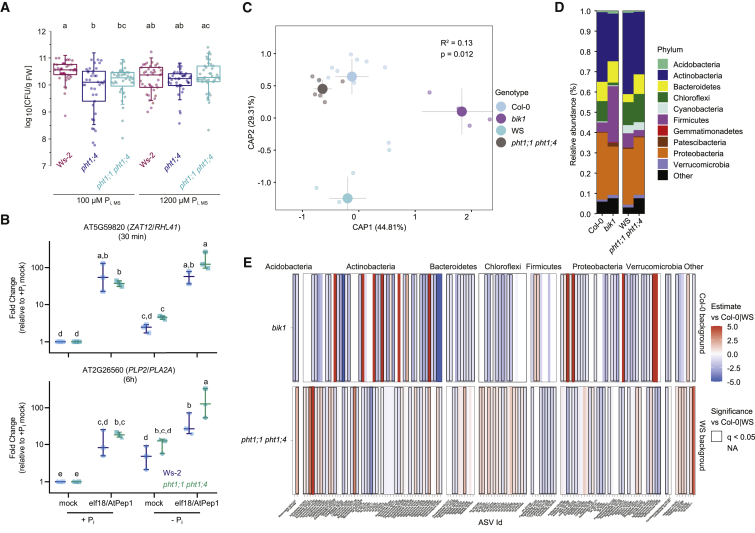
To interrogate the function of PHT1;4 in plant defense responses, we performed infection assays with the soil-borne bacterial pathogen Ralstonia solanacearum GMI1000.42 Inoculation of the root tip and subsequent sampling of shoot tissue revealed that wild-type Ws-2 seedlings are more susceptible to Ralstonia than pht1;4 or pht1;1 pht1;4 seedlings (Figure 4A). Importantly, we observed this PHT1;4-dependent susceptibility only at reduced Pi concentrations in the growth medium, when the abundance of PHT1;4 at the PM should be increased. These results therefore suggest that PHT1;4 and Pi availability normally inhibit plant immunity. This is consistent with the recent findings in rice showing that the PHT1 family member OsPT8 inhibits disease resistance43 and that excess Pi increases susceptibility to pathogens.44 Additionally, elicitor-induced expression of two marker genes (one short-45 and one long-term46) was increased in pht1;1 pht1;4 seedlings grown in Pi-deficient media (Figure 4B), suggesting that limitation of Pi transport can increase immune responses. In keeping with an immune-modulating role of PHT1-mediated Pi transport and previous observations that Pi status can shape the root microbiome,9,10 we observed an altered microbiome structure in pht1;1 pht1;4 mutants, as well as in the bik1 mutant (Figures 4C–4E). Together, these results provide a rationale for the inhibition of Pi transport as a fast PTI response that can shape plant defense responses.
Figure 4.
PHT1-meditated Pi transport shapes immune responses
(A) Ralstonia solanacearum infection assays. Seedlings were grown on the indicated Pi concentrations. Data combine 4 independent replicates. Boxes indicate the 25th and 75th percentiles, the horizontal line shows the median, and error bars represent ± SD. Closed circles show individual measurements. Significance was tested with a one-way ANOVA and a post hoc Tukey test. Equal letters at the top of the panel indicate p > 0.05.
(B) Transcriptional immune responses are increased in pht1;1 pht1;4 seedlings under low Pi. Fold change expression is shown for ZAT12 and PLP2 after short (30 min) and long-term (6 h) treatment with elicitor cocktail (1 μM elf18 and AtPep1), respectively. Shapes indicate independent experiments (n = 2 or 3 per genotype and treatment); lines indicate the median with 95% confidence interval (CI). Statistically significant differences in expression level were calculated via one-way ANOVA followed by pairwise estimated marginal means comparison with Tukey multiplicity adjustment. Equal letters at the top of the panel indicate p > 0.05.
(C–E) Altered root microbiome structure in pht1;1 pht1;4 and bik1 plants.
(C) Canonical analysis of principal component (CAP) of microbiome composition showing the projected microbiome assembly of the mutants pht1;1 pht1;4 and bik1 and their corresponding genetic backgrounds Ws-2 and Col-0 within root fraction (n = 4–12 per genotype; this experiment was repeated twice). Plants were grown in pots filled with a natural soil. Genotypes are represented by colors following the legend on the right. Variance explained (R2) and p values were obtained using PERMANOVA.
(D) Relative abundance profiles of the main bacterial phyla (see legend on the right) in the root microbiomes of the different genotypes Col-0, bik1, Ws-2, and pht1;1 pht1;4.
(E) Heatmaps showing the enrichment of the different amplicon sequence variants (ASVs) in both mutants pht1;1 pht1;4 and bik1 against their corresponding genetic background Ws-2 (Ws-2 background) or Col-0 (Col-0 background). Rectangles outlined in black are ASVs significantly enriched (red) and depleted (blue) with respect to Ws-2 or Col-0 (q < 0.05).
In summary, we have established a method for the direct, real-time measurement of Pi transport in Arabidopsis and subsequently have identified the major Pi uptake transporters PHT1;4 and PHT1;1 as bona fide BIK1/PBL1 targets, providing a mechanistic link through which Pi homeostasis and immune signaling are integrated. Taken together, our results indicate that active limitation of PHT1;4-dependent Pi uptake capacity promotes plant immunity against pathogenic bacteria and affects the microbiome in roots. Future studies will be needed to further characterize how immune signaling regulates other aspects of Pi transport and homeostasis, as well as how this affects interactions with additional soil microbes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GFP-HRP | Santa Cruz | Cat#sc-9996 HRP; RRID: AB_627695 |
| anti-HA-HRP | Roche | Cat#12013819001; RRID: AB_390917 |
| Bacterial and virus strains | ||
| E. coli BL21(DE3) Rosetta pLysS | Sigma | Cat#70956-M |
| Ralstonia solanacearum GMI1000 | Salanoubat et al.42 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 6xHis-MBP-BIK1 | Kadota et al.37 | N/A |
| 6xHis-MBP-BIK1∗ (kinase-dead) | Kadota et al.37 | N/A |
| 6xHis-MBP-PBL1 | Kadota et al.37 | N/A |
| 6xHis-MBP-PBL1∗ (kinase-dead) | Kadota et al.37 | N/A |
| GST | This paper | N/A |
| GST-PHT1;4-loop | This paper | N/A |
| GST-PHT1;4-C terminus | This paper | N/A |
| GST-PHT1;1-loop | This paper | N/A |
| ATP, −32P- 3000Ci/mmol 10mCi/ml EasyTide | Perkin-Elmer | Cat# NEG502A250UC |
| flg22 | SciLight Biotechnology LLC | N/A |
| elf18 | SciLight Biotechnology LLC | N/A |
| Atpep1 | SciLight Biotechnology LLC | N/A |
| RevertAid Reverse Transcriptase | Thermo | Cat#EP0441 |
| PowerUP SYBR Green Master Mix | Thermo | Cat#A25741 |
| ATP, [γ-32P]- 3000Ci/mmol 10mCi/ml EasyTide | Perkin-Elmer | Cat# NEG502A250UC |
| HisPur Cobalt Resin | Thermo | Cat#89965 |
| GST-Bind Resin | Millipore | Cat#70541 |
| Protease Inhibitor Cocktail | Sigma | Cat#P9599 |
| Murashige & Skoog without Phosphate | Caisson | Cat#MSP11-50LT |
| IGEPAL CA-630 | Sigma | Cat#I3021 |
| Tween-20 | Merck | Cat#P1379-25ML |
| MoBio PowerSoil Kit | MOBIO Laboratories; QIAGEN | Cat#12955-4 |
| mPNA | PNA Bio | Cat#MP01-50 |
| pPNA | PNA Bio | Cat#PP01-50 |
| Kapa Robust Taq | Sigma | Cat#KK5518 |
| AMPure XP magnetic beads | Beckman Coulter | Cat#A63881 |
| Kapa HiFi Hotstart Readymix | Roche | Cat#KK2602 |
| Deposited data | ||
| 16 s rRNA sequencing data | This paper | NCBI bio-project: PRJNA771416 |
| Experimental models: organisms/strains | ||
| A. thaliana Ws-2 | N/A | N/A |
| A. thaliana (Ws-2) pht1;4 | Shin et al.3 | N/A |
| A. thaliana (Ws-2) pht1;1 pht1;4 | Shin et al.3 | N/A |
| A. thaliana Col-0 | N/A | N/A |
| A. thaliana (Col-0) bik1 pbl1 | Zhang et al.33 | N/A |
| A. thaliana (Col-0) pht1;4_SALK_138643 | This paper | SALK_138643 |
| A. thaliana (Col-0) pht1;4_SAIL_1225_F08 | This paper | SAIL_1225_F08 |
| A. thaliana (Col-0) pht1;1 | Shin et al.3 | N/A |
| A. thaliana (Ws-2) pht1;1 pht1;4 pPHT1;4::PHT1;4-GFP | This paper | N/A |
| A. thaliana (Col-0) pBIK1::BIK1-HA | Zhang et al.33 | N/A |
| A. thaliana (Col-0) 35S::Lti6b-GFP | Kadota et al.37 | N/A |
| Oligonucleotides | ||
| Primers for cloning and qPCR, see Table S1 | This paper | N/A |
| Primers for bacterial 16S rRNA amplification and sequencing, see Table S2 | This paper. | N/A |
| Recombinant DNA | ||
| pDONRZeo:PHT1;4 | This paper | N/A |
| pGEX4T1:PHT1;4-loop | This paper | N/A |
| pGEX4T1:PHT1;4-C terminus | This paper | N/A |
| 35S::Lti6b-GFP | Kadota et al.37 | N/A |
| 35S::FLS2-GFP | This paper | N/A |
| 35S::PHT1;4-GFP | This paper | N/A |
| pENTR:pPHT1;4::PHT1;4 | Thibaud et al.5 | N/A |
| pPHT1;4::PHT1;4-GFP | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism v8.0.1 | GraphPad | https://www.graphpad.com |
| PatchMaster v2x90-3 | HEKA Electronics | https://heka.com/ |
| Inskscape v0.92.4 | Free Software Foundation, Inc | https://inkscape.org/ |
| R v4.1.1 | R Foundation for Statistical Computing | https://www.r-project.org/ |
| OriginPro 2016 b9.3.1.273 | OriginLab | https://www.originlab.com/origin |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Cyril Zipfel (cyril.zipfel@botinst.uzh.ch).
Materials availability
Materials generated in this study are available from the lead contact without restrictions.
Experimental model and subject details
Arabidopsis mutants and transgenic lines were in Col-0 or Ws-2 background as indicated in Method Details and figure legends. Unless otherwise stated, all seeds were stratified for 2 days in the dark at 4°C before germination.
For in vivo electrophysiology, Arabidopsis seeds were sterilized by treatment with 5% (v/v) NaOCl for 10 min, followed by a treatment with 70% (v/v) EtOH for 10 min, which was followed by 3 to 5 washing steps with sterile MilliQ H2O. Seeds were subsequently placed (three per dish) on the surface of 1 mL sterile ¼ MS-media (5.2 mM NH4NO3; 4.7 mM KNO3; 600 μM CaCl2; 200 μM MgCl2; 25.1 μM H3BO4; 25 μM FeNaEDTA; 19 μM Mn(II)Cl2; 13.3 μM ZnCl2; 1.7 μM KI; 386 nM Na2MoO4; 30 nM CoCl2; 25 nM Cu(II)Cl2; 0.5% (w/v) sucrose; 2.4 mM MES; pH 5.8 with TRIS; 1% (w/v) agarose; KH2PO4 was added to the indicated concentrations and the potassium concentration of the growth medium was balanced with KCl) inside Ø 3.5-cm Petri dishes, which were sealed with parafilm, placed upright in styrofoam cuvette boxes, stratified, and subsequently moved to a KKD Hiros chamber growth chamber (Clitec). Unless otherwise indicated, all electrophysiological measurements were performed on seedlings grown on P0 media (no added phosphate). Germination and seedling growth took place in a 5-day period under 12 h day/12 h night (21°C/16°C, respectively) and a photon flux density of 120 μmol photons m-2 s-1.
For 33Pi uptake experiments, Arabidopsis seeds were surface-sterilized and sown in vitro on square Petri plates with solid modified MS/10 medium47 containing 0.15 mM MgSO4, 2.1 mM NH4NO3, 1.9 mM KNO3, 0.34 mM CaCl2, 0.5 μM KI, 10 μM H3BO3, 10 μM MnSO4, 3 μM ZnSO4, 0.1 μM CuSO4, 0.1 μM CoCl2, 1 μM Na2MoO4, 3.4 mM MES (pH 5.8), 0.5% (w/v) sucrose and 0.8% (w/v) agar. Media was supplemented with 2 μM FeCl2. Then, Pi was added at 0.5 mM KH2PO4 (complete media or +P). After stratification, plants were grown in vitro (16 h day/8 h night at respectively 23°C/21°C) under fluorescent light at 70 to 100 μmol photons m-2 s-1.
For phosphate-dependent elicitor response, surface-sterilized Arabidopsis seeds were sown on ¼ strength MS media plates containing 1% (w/v) sucrose, stratified 3 days, and moved to the growth chamber with conditions 16 h day/8 h night at respectively 22°C/18°C and 120 μmol photons m-2 s-1. At 5 days, seedlings were transferred two-per-well to 24-well plates, with each well containing 1 mL ¼ strength liquid MS media containing 1% (w/v) sucrose. After 5 more days, when seedlings were 10 days old, liquid media was removed, wells were washed with sterile water, and media was replaced with 1 mL either ¼ strength liquid MS 1% (w/v) sucrose, or the same media lacking phosphate (+Pi = 1200 μM, -Pi = 0 μM). After two days, when seedlings were 12 days old, 100 μl of either an elicitor solution in sterile water or a sterile water mock treatment was added to each well. The elicitor solution comprised 10 μM elf18, 10 μM AtPep1, for final concentration of 1 μM each elicitor for treatment. Seedlings were removed from the solution, blot-dried, and flash-frozen in liquid nitrogen at 30 min or 6 h post-treatment.
For coIPs in Arabidopsis seedlings, surface-sterilized Arabidopsis F2 seed (pht1;1 pht1;4 PHT1;4pro:PHT1;4-GFP x Col-0 BIK1pro:BIK1-HA or Col-0 35Spro:Lti6b-GFP x Col-BIK1pro:BIK1-HA) were sown on MS media containing 1% (w/v) sucrose supplemented with 10 μg ml-1 phosphinothricin, stratified, and moved to growth chamber with conditions 16 h day/8 h night at respectively 22°C/18°C and 120 μmol photons m-2 s-1. At 5 days, phosphinothricin-resistant seedlings were transferred five-per-well to 6-well plates, with each well containing 5 mL liquid MS media containing 1% (w/v) sucrose. After 7 more days, when seedlings were 12 days old, liquid media was removed, wells were washed with sterile water, and media was replaced with 5 mL liquid MS 1% (w/v) sucrose lacking phosphate. Seedlings were harvested at 14 days old in liquid nitrogen.
For Ralstonia infection assays, surface-sterilized seeds were sown on ½ MS with 0.5% (w/v) sucrose. After stratification, seeds were germinated in the growth chamber (Percival I-36VL) at 22°C, 16 h light/8 h dark, 100-150 μmol photons m-2 s-1. After 5 days, seedlings were transferred to ½ MS plates containing no sucrose but with the indicated Pi concentrations.
For microbiome analysis, seeds of mutants pht1;1 pht1;4, bik1 and their corresponding backgrounds Ws-2 and Col-0 were surface-sterilized with 70% (v/v) bleach, 0.2% (v/v) Tween-20 for 8 min, and rinsed 3 times with sterile distilled water to eliminate microbes on the seed surface. Seeds were then stratified at 4°C in the dark for 1 day and germinated in sterile pots filled with a natural soil collected from a location free of pesticide and fertilizer at Sutton-Bonington campus, University of Nottingham, UK mixed in a proportion 2:1 (v/v) with autoclaved dry pavior sand. All pots were randomized using the website random.org, and the position of the trays changed every week in the growth chamber (Conviron GEN1000). All pots were watered once a week from the top using sterile distilled water without chlorine and other tap water additives. Plants were grown in a growth chamber under a short-day regime (8h light/16h dark) at 21°C day/19°C night for 11 weeks.
N. benthamiana plants were grown in a greenhouse at a day-night cycle of 16 h/8 h at 25°C/22°C, and a constant 60% rel. humidity.
Method details
In vivo electrophysiology
Hydroponically grown 5-day-old A. thaliana seedlings were covered with ca. 3 mL of bath solution (1 mM CaCl2; 0.1 mM KCl; 5 mM MES; pH 6 with BTP) for at least 20 min prior to measurements. In case seedlings were pre-treated with an elicitor mix, they were first accustomed to the bath solution to which then elicitors were added for at least 15 min. Bulging root hair cells were impaled under microscopic inspection (40x magnification, inverse Nikon Eclipse Ti) with sharp single-barreled micro-electrodes pulled with a DMZ-Universal Puller (Zeitz-Instruments; Germany) from borosilicate glass capillaries (Pr-No:1403547, Hilgenberg, Germany). The electrodes were backfilled with 0.3 M KCl and were connected via a chlorinated silver wire and an electrode holder to a EPC10 USB red star headstage (HEKA electronics, Germany) mounted on a Scientifica PatchStar micromanipulator (Scientifica, UK) A glass capillary filled with 0.3 M KCl and sealed with 0.3 M KCl in 2% (w/v) agarose served as a reference electrode connected via a chlorinated silver wire to the head stage. Signal acquisition was via an EPC10 USB single patch-clamp amplifier with an LIH 8+8 AD/DA interface (HEKA electronics, Germany). under the control of the PatachMaster software (HEKA electronics, Germany). The free running membrane potential was recorded with PatchMaster v2x90-3 Software (HEKA electronics, Germany). The electrode current was clamped to zero and the signal was filtered at 10000 Hz and sampled with 100 Hz. Recordings were started after successful impalement and Pi-supplemented (concentrations as indicated) bath solution was applied via a constantly running gravity perfusion system (flowrate ca. 5 ml/min) after 10 s of stable membrane potential readings. The perfusion outlet was positioned directly above the root near the impaled root hair cell.
RT-qPCR
For PHT1 transcript analysis, total RNA was extracted from 3 to 5 five-day-old seedlings of the indicated genotypes grown on ¼ MS plates without or with 312 μM Pi. Frozen tissue was ground and total RNA was extracted with TRI reagent (Ambion) according to the manufacturer’s instructions. RNA was quantified with a Nanodrop spectrophotometer (Thermo Fischer Scientific). cDNA was synthesized with oligo-dT primer from 460 ng RNA with RevertAid Reverse Transcriptase (Thermo Fischer Scientific) according to the manufacturer’s instructions. Transcripts of PHT1;1, PHT1;4 and ACTIN2 as the reference gene were analyzed through qPCR using PowerUP SYBR Green Master mix (Thermo Fischer Scientific) running on an Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fischer Scientific) using a program of 95°C for 20 s, 40 cycles of 95°C for 3 s and 60°C for 30 s, and the melt curve.
For elicitor response transcript analysis, total RNA was extracted and cDNA prepared as described above, except that 12-day-old seedlings were harvested from the indicated conditions. Fold change relative to full phosphate mock treatment is shown for each genotype, and statistical differences were calculated based on normally-distributed ΔCt values relative to UBOX reference gene, in R48 using tools of the tidyverse49, package emmeans50, and package multcompView51.
For qPCR experiments, a single amplified product was verified through melt curve analysis for each primer pair used, and relative expression was analyzed using the 2(-ΔCt) method. Primers used for qPCR are listed in Table S1.
Co-immunoprecipitation
For transient assays52, leaves of 4-week-old N. benthamiana plants were infiltrated with Agrobacterium tumefaciens (strain GV3101) carrying constructs as indicated in figure captions. In all cases cultures were co-infiltrated with A. tumefaciens carrying a P19 suppressor of gene silencing construct. P19 and Lti6b-GFP constructs were infiltrated at OD600 of 0.1 and 0.05, respectively, and all others at 0.2 in infiltration medium (10 mM MgCl2, 10 mM MES pH 5.7, supplemented with 100 μM acetosyringone). Leaves were detached 2 days post-infiltration and frozen in liquid nitrogen. Tissue (Arabidopsis seedlings or N. benthamiana leaves) was ground in liquid nitrogen and protein extraction and immunoprecipitation were performed as described previously37. Briefly, proteins were extracted from liquid nitrogen-ground tissue in buffer (50 mM Tris-Cl pH 7.5, 150 nM NaCl, 10% (v/v) glycerol, 2 mM EDTA, 5 mM DTT, 1% (v/v) IGEPAL, supplemented with protease inhibitor cocktail P9599 (Sigma)) and immunoprecipitation was performed with GFP-clamp resin53 at 4°C. Bound proteins were washed three times in buffer and were eluted by heating in SDS loading-dye for 10 min at 80°C. Proteins were separated by SDS-PAGE and blotted onto PVDF membrane. Proteins were blocked and probed in TBST-5% (w/v) non-fat milk using anti-GFP (HRP-conjugated B-2, sc-9996 HRP, Santa Cruz, 1:5000 dilution) or anti-HA (HRP-conjugated, 12013819001, Roche, 1:3000 dilution).
Molecular cloning
The CDS of PHT1;4 was cloned into pDONRZeo using BP Clonase II (Invitrogen) according to manufacturer instructions. For expression as GFP-fusions in N. benthamiana, PHT1;4 or FLS2 were subcloned into the pGWB605 destination vector54 using LR Clonase II (Invitrogen). The cytosolic loop (aa 234-294) and C terminus (aa 503-534) of PHT1;4 and the cytosolic loop of PHT1;1 (aa 233-293) were each cloned into SalI/NotI-digested pGEX4T1 using the primers provided (Table S1). For native promoter PHT1;4-GFP lines, PHT1;4pro:PHT1;45 was cloned from pENTR into the pGWB601 destination vector54 using LR-II Clonase (Invitrogen).
Recombinant protein expression and purification
All recombinant proteins used in this study were expressed in Escherichia coli strain BL21(DE3) Rosetta pLysS. BIK1, BIK1∗ (kinase-dead, K105A/K106A), PBL1, and PBL1∗ (kinase-dead, K99E) were all expressed as 6xHis-MBP fusions using the pOPINM vector. Recombinant proteins were affinity purified using HisPur Cobalt Resin (Thermo) or GSH Sepharose (GE Healthcare).
In vitro kinase assays
Approximately 1 μg of kinase was incubated with approximately 2 μg of substrate protein in kinase buffer (25 mM Tris-Cl pH 7.5, 5 mM MgCl2, 5 mM MnCl2, 1 mM DTT), and reactions were performed at RT for 30 min with the addition of 5 μM ATP + 0.5 μCi 32P-γ-ATP. Autoradiographs were imaged following SDS-PAGE and transferred to PVDF membrane using an Amersham Typhoon phosphorimager (GE Healthcare).
33P uptake measurements
Seven-day-old seedlings were transferred in vitro to media with or without 100 nM elicitor for 5 days. An additional 250 μL of 100 nM sterile elicitor solution was provided to the root system of each plantlet daily. To measure Pi uptake, each root system was immersed in 4 mL incubation medium (0.1 mM CaCl2, 5 mM MES, 50 μM KH2PO4 pH 5.5, 0.15 μCi 33Pi/ml) for 2 h at room temperature (22-24°C) under white light (150-180 μE m-2 s-1). Seedlings were rinsed and immersed in 3 mL desorption solution (0.1 mM CaCl2, 5 mM MES, 1 mM KH2PO4, pH 5.5) for 2 h on ice. Plantlets were dried and radioactivity measurements are performed with a beta counter27.
Ralstonia infection assays
8-day-old seedlings were inoculated by placing 5 μL of a bacterial inoculum containing 105 CFU/mL of R. solanacearum GMI1000 on the root (0.5-1 cm to the root tip) of each seedling55. Samples of shoot tissue were taken and processed 3 days post-inoculation. Shoots were weighed, ground in distilled water, and serial dilutions were plated on solid BG medium to quantify bacterial numbers as CFU/g of fresh weight.
Microbiome analyses
After 11 weeks, roots were harvested separately from the individual pots and placed in 50 mL Falcon tubes containing sterile water. Root samples were then rinsed three times with sterile distilled water to remove weakly associated microbes and soil particles and stored at −80°C until further analysis. For DNA extraction we used 96-well-format MoBio PowerSoil Kit (MOBIO Laboratories; QIAGEN) following the manufacturer’s instruction. Root samples were lyophilized using an Alpha 2-4 LD freeze dry system, and pulverized using a TissueLyser II (QIAGEN), 2 cycles of 30 s, frequency 30 s-1. Before starting the extraction, all samples, were randomized mechanically using a plastic bag. Samples were then taken individually from the bag and loaded in the DNA extraction plates.
For 16S rRNA sequencing, the V3-V4 region of the bacterial 16S rRNA gene was amplified using the primers 338F and 806R. Two barcodes and six frameshifts were added to the 5′ end of 338F and six frameshifts were added to the 806R primers. PCR reactions with ∼20 ng DNA template were performed in triplicate and included a unique mixture of three frameshifted primer combinations for each plate. PCR blockers mPNA and pPNA were used to reduce contamination by plant host plastid and mitochondrial 16S rRNA amplicon. PCR conditions were as follows: 5 μL Kapa Enhancer, 5 μL Kapa Buffer A, 1.25 μL 5 μM 338F, 1.25 μL 5 μM 806R, 0.375 μL mixed plant rRNA gene-blocking peptide nucleic acids (PNAs; 1:1 mix of 100 μM plastid PNA and 100 μM mitochondrial PNA), 0.5 μL Kapa dNTPs, 0.2 μL Kapa Robust Taq, 8 μL dH2O, 5 μL DNA; temperature cycling: 95°C for 60 s; 24 cycles of 95°C for 15 s; 78°C (PNA) for 10 s; 50°C for 30 s; 72°C for 30 s; 12°C until use.
PCR reactions were clean-up using AMPure XP magnetic beads (Beckman Coulter) to remove primer dimers. The PCR product was indexed using a primer that contains forward Illumina adaptor (Table S1) and 96 indexed reverse primers (5′-CAAGCAGAAGACGGCATACGAGATXXXXXXXXXGTGACTGGAGTTCAGACGTGTGCTC-3′) with reverse Illumina adaptor (Table S2). We used Kapa HiFi Hotstart Readymix and the following temperature cycling: 95°C for 60 s; 9 cycles of 95°C for 15 s; 78°C (PNA) for 10 s; 60°C for 30 s; 72°C for 35 s; 12°C until use.
PCR products were purified using AMPure XP magnetic beads (Beckman Coulter) and quantified with a Qubit 2.0 fluorometer (Invitrogen). Amplicons were pooled in equal amounts and then diluted to 10 pM for sequencing. Sequencing was performed on an Illumina MiSeq instrument using a 600-cycle V3 chemistry kit at the DeepSeq facility at the University of Nottingham. The abundance matrix, metadata and taxonomy are available at https://github.com/isaisg/Pht1-BIK.
Sequences data were processed with MT-Toolbox56. The resulting sequences were then denoised and collapsed into amplicon sequence variants (ASVs), using DADA2 v.1.10.157. Representative ASVs sequences were taxonomically classified with the mothur naive bayes classifier58 trained on the SILVA 132 database59. ASVs, with more than 1000 reads per sample, were used to create the raw count abundance tables. The resulting abundance tables were processed and analyzed with functions from the ohchibi package (https://github.com/isaisg/ohchibi).
Beta diversity analyses (canonical analysis of principal coordinates) were based on Bray-Curtis dissimilarity matrices calculated from the abundance table. We used the capscale function from the vegan R package v.2.5-559 to compute the canonical analysis of principal coordinates (CAP), with the following design:
BrayCurtis Dissimilarity ∼Genotype + Condition (Rep)
We used the R packages MASS, pscl and stats to compute the mutants specific enrichment profiles independently. For each ASV, we estimated their abundance differences in each mutant against its background, using a set of generalized linear models (GLM) with the following design:
Abundance ∼Genotype + Rep
We picked the GLM that best fit each individual ASV via the Akaike information criterion (AIC). We adjusted the global p-value table using the False Discovery Rate (FDR) approach. In a given mutant, an ASV with a corrected p-value < 0.05 was considered differentially abundant against Col-0 or Ws-2 in each case. We created a heatmap in which we visualized the enrichment patterns (estimate with respect to Col-0 or Ws-2) of all statistically significant ASV for both mutants used using ggplot2 v.3.2.1 package60 in R. The experiments were repeated twice.
Quantification and statistical analysis
Data were visualized using GraphPad Prism, R, or Origin as described in Method details. Statistical tests, n values, and significance cutoffs are described in figure captions and/or Method details.
Acknowledgments
This work was supported by the European Research Council under the European Union (EU)’s Horizon 2020 research and innovation program (grant agreements no. 309858, project “PHOSPHinnATE,” and no. 773153, project “IMMUNO-PEPTALK,” to C.Z.), The Gatsby Charitable Foundation (to C.Z.), the University of Zürich (to C.Z.), and the Swiss National Science Foundation (grant 31003A_182625 to C.Z.). J.D. and T.A.D. were supported by postdoctoral fellowships from the European Molecular Biology Organization (EMBO LTFs no. 683-2018 to J.D. and no. 100-2017 to T.A.D.). T.A.D. was further supported by the Natural Sciences and Engineering Research Council of Canada (NSERC PDF-532561-2019). V.C. and G.C. were supported by the Biotechnology and Biological Sciences Research Council and National Science Foundation (BBSRC-NSF) (grant no. BB/V011294/1 to G.C.). G.Y. and L.Z. were supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB27040204 to A.P.M.), the Chinese 1000 Talents Program (to A.P.M.), and the Shanghai Center for Plant Stress Biology. The authors thank all members of the Zipfel lab for fruitful discussion and feedback on the manuscript.
Author contributions
J.D., T.A.D., and C.Z. conceived and designed the project. J.D., T.A.D., G.Y., L.Z., P.D., M.B., M.-C.T., V.C., and G.C. generated materials, performed experiments, and/or analyzed data. L.N., A.P.M., and C.Z. supervised the project. J.D., T.A.D., and C.Z. wrote the manuscript with input from all authors.
Declaration of interests
The authors declare no competing interests.
Published: December 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.11.063.
Supplemental information
Data and code availability
-
•
16 s rRNA sequencing data have been deposited at the NCBI bio-project and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. Additional data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Bieleski R.L. Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 1973;24:225–252. [Google Scholar]
- 2.Nussaume L., Kanno S., Javot H., Marin E., Pochon N., Ayadi A., Nakanishi T.M., Thibaud M.C. Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2011;2:83. doi: 10.3389/fpls.2011.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin H., Shin H.-S., Dewbre G.R., Harrison M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004;39:629–642. doi: 10.1111/j.1365-313X.2004.02161.x. [DOI] [PubMed] [Google Scholar]
- 4.Misson J., Thibaud M.C., Bechtold N., Raghothama K., Nussaume L. Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 2004;55:727–741. doi: 10.1007/s11103-004-1965-5. [DOI] [PubMed] [Google Scholar]
- 5.Thibaud M.C., Arrighi J.F., Bayle V., Chiarenza S., Creff A., Bustos R., Paz-Ares J., Poirier Y., Nussaume L. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 2010;64:775–789. doi: 10.1111/j.1365-313X.2010.04375.x. [DOI] [PubMed] [Google Scholar]
- 6.Bayle V., Arrighi J.-F.F., Creff A., Nespoulous C., Vialaret J., Rossignol M., Gonzalez E., Paz-Ares J., Nussaume L. Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell. 2011;23:1523–1535. doi: 10.1105/tpc.110.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park B.S., Seo J.S., Chua N.H. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell. 2014;26:454–464. doi: 10.1105/tpc.113.120311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao Y.Y., Li J.L., Pan R.L., Chiou T.J. Structure-function analysis reveals amino acid residues of Arabidopsis phosphate transporter AtPHT1;1 crucial for its activity. Front. Plant Sci. 2019;10:1158. doi: 10.3389/fpls.2019.01158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castrillo G., Teixeira P.J.P.L., Paredes S.H., Law T.F.T.F., de Lorenzo L., Feltcher M.E., Finkel O.M., Breakfield N.W., Mieczkowski P., Jones C.D., et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkel O.M., Salas-González I., Castrillo G., Spaepen S., Law T.F., Teixeira P.J.P.L., Jones C.D., Dangl J.L. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol. 2019;17:e3000534. doi: 10.1371/journal.pbio.3000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J., Zhao B., Zheng S., Zhang X., Wang X., Dong W., Xie Q., Wang G., Xiao Y., Chen F., et al. A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell. 2021;184:5527–5540.e18. doi: 10.1016/j.cell.2021.09.030. [DOI] [PubMed] [Google Scholar]
- 12.Ullrich-Eberius C.I., Novacky A., Fischer E., Lüttge U. Relationship between energy-dependent phosphate uptake and the electrical membrane potential in Lemna gibba G1. Plant Physiol. 1981;67:797–801. doi: 10.1104/pp.67.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ullrich-Eberius C.I., Novacky A., van Bel A.J. Phosphate uptake in Lemna gibba G1: energetics and kinetics. Planta. 1984;161:46–52. doi: 10.1007/BF00951459. [DOI] [PubMed] [Google Scholar]
- 14.Dindas J., Scherzer S., Roelfsema M.R.G., von Meyer K., Müller H.M., Al-Rasheid K.A.S., Palme K., Dietrich P., Becker D., Bennett M.J., Hedrich R. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat. Commun. 2018;9:1174. doi: 10.1038/s41467-018-03582-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raghothama K.G. Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- 16.Abel S. Phosphate scouting by root tips. Curr. Opin. Plant Biol. 2017;39:168–177. doi: 10.1016/j.pbi.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Hamburger D., Rezzonico E., MacDonald-Comber Petétot J., Somerville C., Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 2002;14:889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv Q., Zhong Y., Wang Y., Wang Z., Zhang L., Shi J., Wu Z., Liu Y., Mao C., Yi K., Wu P. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell. 2014;26:1586–1597. doi: 10.1105/tpc.114.123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z., Ruan W., Shi J., Zhang L., Xiang D., Yang C., Li C., Wu Z., Liu Y., Yu Y., et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. USA. 2014;111:14953–14958. doi: 10.1073/pnas.1404680111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puga M.I., Mateos I., Charukesi R., Wang Z., Franco-Zorrilla J.M., de Lorenzo L., Irigoyen M.L., Masiero S., Bustos R., Rodríguez J., et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:14947–14952. doi: 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wild R., Gerasimaite R., Jung J.Y., Truffault V., Pavlovic I., Schmidt A., Saiardi A., Jessen H.J., Poirier Y., Hothorn M., Mayer A. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science. 2016;352:986–990. doi: 10.1126/science.aad9858. [DOI] [PubMed] [Google Scholar]
- 22.Wege S., Khan G.A., Jung J.Y., Vogiatzaki E., Pradervand S., Aller I., Meyer A.J., Poirier Y. The EXS domain of PHO1 participates in the response of shoots to phosphate deficiency via a root-to-shoot signal. Plant Physiol. 2016;170:385–400. doi: 10.1104/pp.15.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ried M.K., Wild R., Zhu J., Pipercevic J., Sturm K., Broger L., Harmel R.K., Abriata L.A., Hothorn L.A., Fiedler D., et al. Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. Nat. Commun. 2021;12:384. doi: 10.1038/s41467-020-20681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., Fu S., Yang L., Luan M., Zhao F., Luan S., Lan W. Vacuolar SPX-MFS transporters are essential for phosphate adaptation in plants. Plant Signal. Behav. 2016;11:e1213474. doi: 10.1080/15592324.2016.1213474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J., Yang L., Luan M., Wang Y., Zhang C., Zhang B., Shi J., Zhao F.G., Lan W., Luan S. A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2015;112:E6571–E6578. doi: 10.1073/pnas.1514598112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunlop J., Phung H.T., Meeking R., White D.W.R. The kinetics associated with phosphate absorption by Arabidopsis and its regulation by phosphorus status. Aust. J. Plant Physiol. 1997;24:623–629. [Google Scholar]
- 27.Ayadi A., David P., Arrighi J.F., Chiarenza S., Thibaud M.C., Nussaume L., Marin E. Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol. 2015;167:1511–1526. doi: 10.1104/pp.114.252338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith S.E., Jakobsen I., Grønlund M., Smith F.A. Roles of arbuscular mycorrhizas in plant phosphorus nutrition: interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol. 2011;156:1050–1057. doi: 10.1104/pp.111.174581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hacquard S., Kracher B., Hiruma K., Münch P.C., Garrido-Oter R., Thon M.R., Weimann A., Damm U., Dallery J.F., Hainaut M., et al. Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 2016;7:11362. doi: 10.1038/ncomms11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 31.Couto D., Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016;16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 32.Liang X., Zhou J.M. Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu. Rev. Plant Biol. 2018;69:267–299. doi: 10.1146/annurev-arplant-042817-040540. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J., Li W., Xiang T., Liu Z., Laluk K., Ding X., Zou Y., Gao M., Zhang X., Chen S., et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA. 2010;107:496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C., Zhou J.M. A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature. 2012;485:114–118. doi: 10.1038/nature10962. [DOI] [PubMed] [Google Scholar]
- 36.Liu Z., Wu Y., Yang F., Zhang Y., Chen S., Xie Q., Tian X., Zhou J.M. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA. 2013;110:6205–6210. doi: 10.1073/pnas.1215543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D., Shirasu K., Menke F., Jones A., Zipfel C. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 38.Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 2014;15:329–338. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Thor K., Jiang S., Michard E., George J., Scherzer S., Huang S., Dindas J., Derbyshire P., Leitão N., DeFalco T.A., et al. The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature. 2020;585:569–573. doi: 10.1038/s41586-020-2702-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian W., Hou C., Ren Z., Wang C., Zhao F., Dahlbeck D., Hu S., Zhang L., Niu Q., Li L., et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature. 2019;572:131–135. doi: 10.1038/s41586-019-1413-y. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H., Huang L., Hong Y., Song F. BOTRYTIS-INDUCED KINASE1, a plasma membrane-localized receptor-like protein kinase, is a negative regulator of phosphate homeostasis in Arabidopsis thaliana. BMC Plant Biol. 2016;16:152. doi: 10.1186/s12870-016-0841-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salanoubat M., Genin S., Artiguenave F., Gouzy J., Mangenot S., Arlat M., Billault A., Brottier P., Camus J.C., Cattolico L., et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- 43.Dong Z., Li W., Liu J., Li L., Pan S., Liu S., Gao J., Liu L., Liu X., Wang G.L., Dai L. The rice phosphate transporter protein OsPT8 regulates disease resistance and plant growth. Sci. Rep. 2019;9:5408. doi: 10.1038/s41598-019-41718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campos-Soriano L., Bundó M., Bach-Pages M., Chiang S.F., Chiou T.J., San Segundo B. Phosphate excess increases susceptibility to pathogen infection in rice. Mol. Plant Pathol. 2020;21:555–570. doi: 10.1111/mpp.12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bjornson M., Pimprikar P., Nürnberger T., Zipfel C. The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat. Plants. 2021;7:579–586. doi: 10.1038/s41477-021-00874-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stringlis I.A., Proietti S., Hickman R., Van Verk M.C., Zamioudis C., Pieterse C.M.J. Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 2018;93:166–180. doi: 10.1111/tpj.13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnaud C., Clément M., Thibaud M.C., Javot H., Chiarenza S., Delannoy E., Revol J., Soreau P., Balzergue S., Block M.A., et al. Identification of phosphatin, a drug alleviating phosphate starvation responses in Arabidopsis. Plant Physiol. 2014;166:1479–1491. doi: 10.1104/pp.114.248112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.R Development Core Team . 2021. R: A language and environment for statistical computing (R Foundation for Statistical Computing) [Google Scholar]
- 49.Wickham H., Averick M., Bryan J., Chang W., McGowan L.D., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the Tidyverse. J. Open Source Softw. 2019;4:1686. [Google Scholar]
- 50.Lenth R.V. 2021. emmeans: estimated marginal means, aka least-squares means. R package version 1.6.1.https://rdocumentation.org/packages/emmeans/versions/1.6.1 [Google Scholar]
- 51.Hothorn T., Bretz F., Westfall P. Simultaneous inference in general parametric models. Biom. J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y., Dorey S., Swiderski M., Jones J.D.G. Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 2004;40:213–224. doi: 10.1111/j.1365-313X.2004.02201.x. [DOI] [PubMed] [Google Scholar]
- 53.Hansen S., Stüber J.C., Ernst P., Koch A., Bojar D., Batyuk A., Plückthun A. Design and applications of a clamp for green fluorescent protein with picomolar affinity. Sci. Rep. 2017;7:16292. doi: 10.1038/s41598-017-15711-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamura S., Mano S., Tanaka Y., Ohnishi M., Nakamori C., Araki M., Niwa T., Nishimura M., Kaminaka H., Nakagawa T., et al. Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci. Biotechnol. Biochem. 2010;74:1315–1319. doi: 10.1271/bbb.100184. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y., Zhao A., Morcillo R.J.L., Yu G., Xue H., Rufian J.S., Sang Y., Macho A.P. A bacterial effector protein uncovers a plant metabolic pathway involved in tolerance to bacterial wilt disease. Mol. Plant. 2021;14:1281–1296. doi: 10.1016/j.molp.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 56.Yourstone S.M., Lundberg D.S., Dangl J.L., Jones C.D. MT-Toolbox: improved amplicon sequencing using molecule tags. BMC Bioinformatics. 2014;15:284. doi: 10.1186/1471-2105-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickham H. Springer International; 2016. ggplot2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
16 s rRNA sequencing data have been deposited at the NCBI bio-project and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. Additional data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.