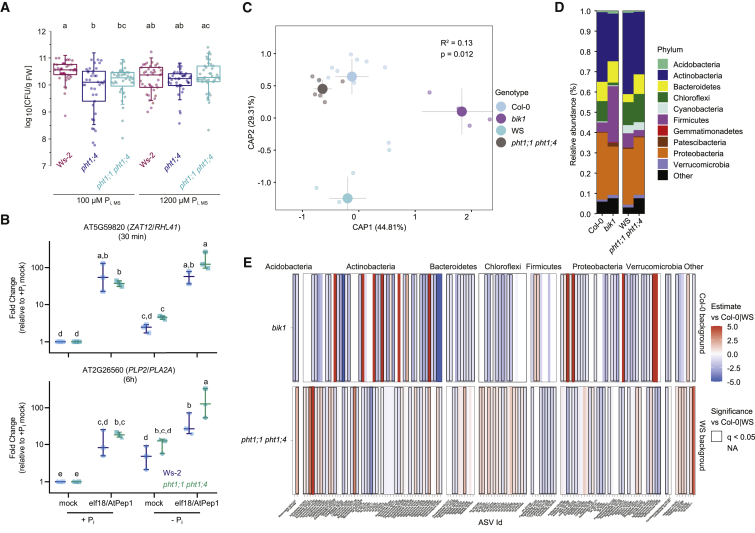
Figure 4.
PHT1-meditated Pi transport shapes immune responses
(A) Ralstonia solanacearum infection assays. Seedlings were grown on the indicated Pi concentrations. Data combine 4 independent replicates. Boxes indicate the 25th and 75th percentiles, the horizontal line shows the median, and error bars represent ± SD. Closed circles show individual measurements. Significance was tested with a one-way ANOVA and a post hoc Tukey test. Equal letters at the top of the panel indicate p > 0.05.
(B) Transcriptional immune responses are increased in pht1;1 pht1;4 seedlings under low Pi. Fold change expression is shown for ZAT12 and PLP2 after short (30 min) and long-term (6 h) treatment with elicitor cocktail (1 μM elf18 and AtPep1), respectively. Shapes indicate independent experiments (n = 2 or 3 per genotype and treatment); lines indicate the median with 95% confidence interval (CI). Statistically significant differences in expression level were calculated via one-way ANOVA followed by pairwise estimated marginal means comparison with Tukey multiplicity adjustment. Equal letters at the top of the panel indicate p > 0.05.
(C–E) Altered root microbiome structure in pht1;1 pht1;4 and bik1 plants.
(C) Canonical analysis of principal component (CAP) of microbiome composition showing the projected microbiome assembly of the mutants pht1;1 pht1;4 and bik1 and their corresponding genetic backgrounds Ws-2 and Col-0 within root fraction (n = 4–12 per genotype; this experiment was repeated twice). Plants were grown in pots filled with a natural soil. Genotypes are represented by colors following the legend on the right. Variance explained (R2) and p values were obtained using PERMANOVA.
(D) Relative abundance profiles of the main bacterial phyla (see legend on the right) in the root microbiomes of the different genotypes Col-0, bik1, Ws-2, and pht1;1 pht1;4.
(E) Heatmaps showing the enrichment of the different amplicon sequence variants (ASVs) in both mutants pht1;1 pht1;4 and bik1 against their corresponding genetic background Ws-2 (Ws-2 background) or Col-0 (Col-0 background). Rectangles outlined in black are ASVs significantly enriched (red) and depleted (blue) with respect to Ws-2 or Col-0 (q < 0.05).